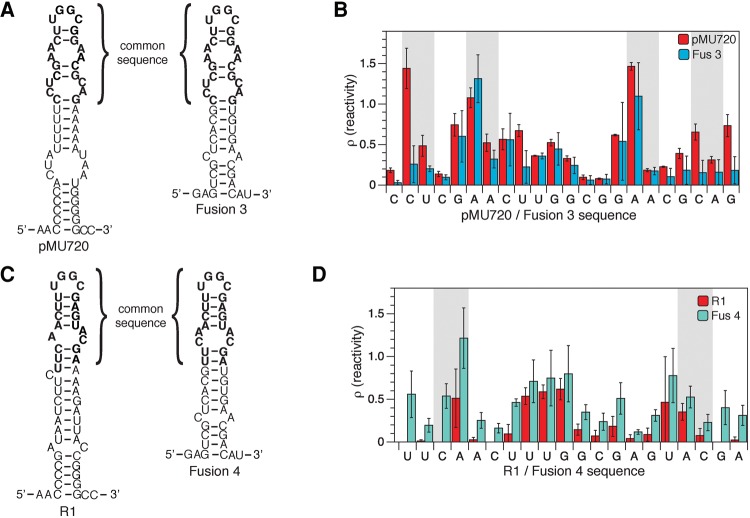
FIGURE 5.
Hairpins from natural kissing hairpin translational regulators have similar nucleotide reactivity patterns as functional chimeric attenuator hairpins. (A) In-cell SHAPE-restrained secondary structure prediction of hairpins from the pMU720 regulator and the Fusion 3 chimeric attenuator. Bold nucleotides indicate the sequence from pMU720 used to create the Fusion 3 chimeric attenuator. (B) In-cell SHAPE-Seq reactivity spectra comparing pMU720 and Fusion 3 for their common nucleotides. Reactivity spectra represent an average of three independent in-cell SHAPE-Seq experiments with error bars representing standard deviations at each nucleotide. (C,D) As in A,B but for hairpins from the R1 regulator and the Fusion 4 chimeric attenuator. Bold nucleotides indicate the sequence from R1 used to create the Fusion 4 chimeric attenuator. Full reactivity spectra can be found in Supplemental Figure S11.