Abstract
Background
Intratumor heterogeneity may foster tumor evolution and adaptation and hinder personalized-medicine strategies that depend on results from single tumor-biopsy samples.
Methods
To examine intratumor heterogeneity, we performed exome sequencing, chromosome aberration analysis, and ploidy profiling on multiple spatially separated samples obtained from primary renal carcinomas and associated metastatic sites. We characterized the consequences of intratumor heterogeneity using immunohistochemical analysis, mutation functional analysis, and profiling of messenger RNA expression.
Results
Phylogenetic reconstruction revealed branched evolutionary tumor growth, with 63 to 69% of all somatic mutations not detectable across every tumor region. Intratumor heterogeneity was observed for a mutation within an autoinhibitory domain of the mammalian target of rapamycin (mTOR) kinase, correlating with S6 and 4EBP phosphorylation in vivo and constitutive activation of mTOR kinase activity in vitro. Mutational intratumor heterogeneity was seen for multiple tumor-suppressor genes converging on loss of function; SETD2, PTEN, and KDM5C underwent multiple distinct and spatially separated inactivating mutations within a single tumor, suggesting convergent phenotypic evolution. Gene-expression signatures of good and poor prognosis were detected in different regions of the same tumor. Allelic composition and ploidy profiling analysis revealed extensive intratumor heterogeneity, with 26 of 30 tumor samples from four tumors harboring divergent allelic-imbalance profiles and with ploidy heterogeneity in two of four tumors.
Conclusions
Intratumor heterogeneity can lead to underestimation of the tumor genomics landscape portrayed from single tumor-biopsy samples and may present major challenges to personalized-medicine and biomarker development. Intratumor heterogeneity, associated with heterogeneous protein function, may foster tumor adaptation and therapeutic failure through Darwinian selection. (Funded by the Medical Research Council and others.)
Large-scale sequencing analyses of solid cancers have identified extensive heterogeneity between individual tumors.1–6 Genetic intratumor heterogeneity has also been shown7–15 and can contribute to treatment failure and drug resistance. Intratumor heterogeneity may have important consequences for personalized-medicine approaches that commonly rely on single tumorbiopsy samples to portray tumor mutational landscapes. Studies comparing mutational profiles of primary tumors and associated metastatic lesions16,17 or local recurrences18 have provided evidence of intratumor heterogeneity at nucleotide resolution. Intratumor heterogeneity within primary tumors and associated metastatic sites has not been systematically characterized by next-generation sequencing. We applied exome sequencing, chromosome aberration analysis, and DNA ploidy profiling to study multiple spatially separated biopsy samples from primary renal-cell carcinomas and associated metastatic sites. We investigated the phenotypic consequences of genetic intratumor heterogeneity and the representation of the tumor genomic landscape by a single tumorbiopsy sample, the current basis for most biomarker discovery and personalized-medicine approaches.
Methods
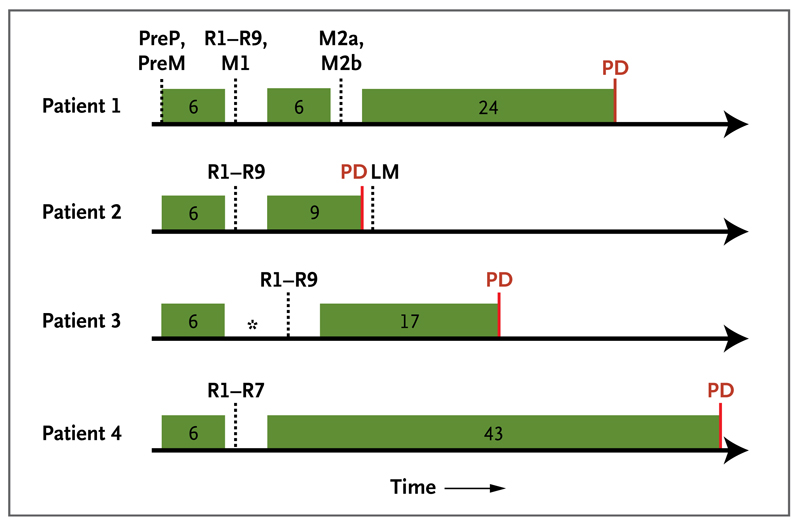
We evaluated tumor-biopsy samples from four consecutive patients with metastatic renal-cell carcinoma who were enrolled in the Personalized RNA Interference to Enhance the Delivery of Individualized Cytotoxic and Targeted Therapeutics clinical trial of everolimus (E-PREDICT; EudraCT number, 2009-013381-54) before and after cytoreductive nephrectomy. Biopsy samples were obtained before the initiation of 6 weeks of treatment with everolimus. After a 1-week washout period in which patients did not receive everolimus, a nephrectomy was performed. Everolimus treatment was continued after recovery from surgery until tumor progression. Figure 1 shows biopsy and treatment timelines.
Figure 1. Biopsy and Treatment Timelines for the Four Patients.
Exon-capture sequencing was performed on tumor DNA from pretreatment biopsy samples of the primary tumor (PreP) and chest-wall metastasis (PreM), primary-tumor regions of the nephrectomy specimen (R1 to R9), a perinephric metastasis in the nephrectomy specimen (M1), and two regions of the excised chest-wall metastasis (M2a and M2b). LM denotes liver metastasis, and PD progressive disease. Green boxes indicate periods of everolimus treatment, with the treatment duration provided in weeks. Dotted lines indicate time points of biopsies, and the asterisk indicates a delay in nephrectomy because of toxicity.
We performed whole-exome multiregion spatial sequencing on DNA that was extracted from freshfrozen samples obtained from Patients 1 and 2, as described previously,19 with paired-end reads of 72 bp and 75 bp, respectively, on Illumina Genome Analyzer IIx and HiSeq platforms. We performed single-nucleotide polymorphism (SNP) array analysis on Illumina Omni2.5 and messenger RNA (mRNA) expression profiling on Affymetrix Gene 1.0 arrays.
All four patients provided written informed consent. Details regarding materials and methods are provided in the Supplementary Appendix, available with the full text of this article at NEJM.org. The study protocol is also available at NEJM.org.
Results
Patients
Patient 1 had a clear-cell carcinoma, pulmonary metastases, and a chest-wall metastasis. Sequencing detected a 2-bp deletion in the von Hippel–Lindau tumor-suppressor gene (VHL) leading to mutational inactivation, which is characteristic of clear-cell carcinoma. After 6 weeks of everolimus treatment and a 1-week washout period, a nephrectomy was performed. The patient restarted everolimus for 6 weeks and after another 1-week washout period proceeded to surgery of the chest-wall mass (Fig. 1). Computed tomography (CT) did not reveal any change in the dimensions of the primary tumor or chest-wall metastasis during everolimus treatment.
Identification and Validation of Somatic Mutations
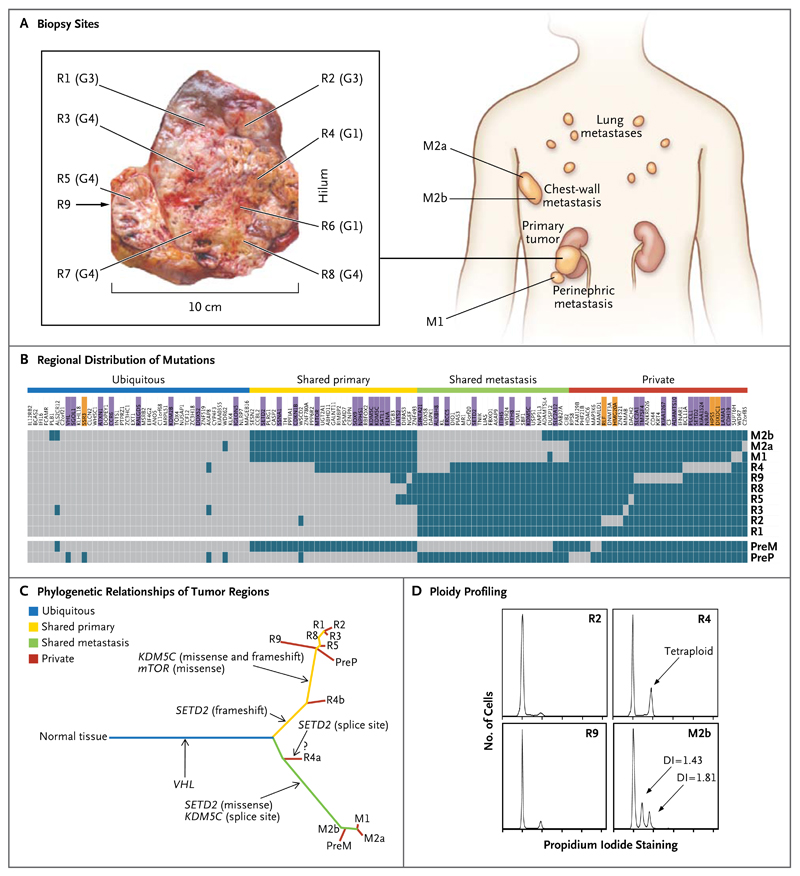
For Patient 1, we performed exon-capture multiregion sequencing on DNA from pretreatment biopsy samples of the primary tumor (PreP) and chestwall metastasis (PreM), nine primary-tumor regions of the nephrectomy specimen (R1 to R9), a metastasis in the perinephric fat of the nephrectomy specimen (M1), two regions of the excised chestwall metastasis (M2a and M2b), and germline DNA19 (Fig. 2A). This sequencing resulted in a median coverage of 74 reads (Table 1 in the Supplementary Appendix). Nonsynonymous somatic point mutations and insertions and deletions (indels) that change the protein amino acid sequence were filtered and manually reviewed to remove sequencing and alignment errors and to determine the regional distribution of mutations. Regions R6 and R7 were excluded from analyses since only one nonsynonymous variant passed filtering. We identified 101 nonsynonymous point mutations and 32 indels (Table 2 in the Supplementary Appendix) and mapped their regional distributions across the tumor (Fig. 2B). Sanger sequencing was used to validate 42 mutations. Of these mutations, 37 (88%) were specifically validated in regions in which they were initially identified (Fig. 2B, andFig. 1 in the Supplementary Appendix), documenting genetic intratumor heterogeneity.
Figure 2. Genetic Intratumor Heterogeneity and Phylogeny in Patient 1.
Panel A shows sites of core biopsies and regions harvested from nephrectomy and metastasectomy specimens. G indicates tumor grade. Panel B shows the regional distribution of 101 nonsynonymous point mutations and 32 indels in seven primary-tumor regions of the nephrectomy specimen (R1 through R5 and R8 through R9), in the perinephric fat of the nephrectomy specimen (M1), and in two regions of the excised chestwall metastasis (M2a and M2b), as detected by exome sequencing (including the VHL mutation detected by Sanger sequencing). Regions R6 and R7 were excluded from analyses since only one nonsynonymous variant passed filtering. The heat map indicates the presence of a mutation (gray) or its absence (dark blue) in each region. The color bars above the heat map indicate classification of mutations according to whether they are ubiquitous, shared by primary-tumor regions, shared by metastatic sites, or unique to the region (private). Among the gene names, purple indicates that the mutation was validated, and orange indicates that the validation of the mutation failed. Because of limited DNA availability, only six mutations were validated in pretreatment samples of the primary tumor (PreP) and chest-wall metastases (PreM) (in VHL, MTOR, SOX9, ALKBH8, SETD2, and KDM5C splice sites). Panel C shows phylogenetic relationships of the tumor regions. R4a and R4b are the subclones detected in R4. A question mark indicates that the detected SETD2 splice-site mutation probably resides in R4a, whereas R4b most likely shares the SETD2 frameshift mutation also found in other primary-tumor regions. Branch lengths are proportional to the number of nonsynonymous mutations separating the branching points. Potential driver mutations were acquired by the indicated genes in the branch (arrows). Panel D shows regional ploidy profiling analysis. All other primary-tumor regions were diploid (not shown). DI denotes DNA index of the aneuploid peak, indicating the DNA content as compared with a diploid genome.
A low false negative mutation call rate is required to avoid overestimation of intratumor heterogeneity. We performed ultradeep exon-capture sequencing of R4 and R9 (median coverage of 262 and 255 reads, respectively) to investigate whether heterogeneous mutations that were not found in R4 or R9 could be detected by increasing the sequencing depth (i.e., the median number of sequencing reads across each exon). This identified all 64 mutations known to be present in R4 and 75 mutations in R9 and detected only 2 additional mutations (in ITGB3 and AKAP8, both in R4) present in other primary regions, indicating a low false negative rate of 2 in 141 (1.4%) (Table 3 in the Supplementary Appendix).
Somatic Mutational Heterogeneity and Clonal Ordering
We excluded 5 mutations that were not validated and classified the remaining 128 mutations into 40 ubiquitous mutations, 59 mutations shared by several but not all regions, and 29 mutations that were unique to specific regions (so-called private mutations) that were present in a single region. We subdivided shared mutations into 31 mutations shared by most of the primary tumor regions of the nephrectomy specimen (R1 to R3, R5, and R8 to R9), pretreatment biopsy samples of the primary tumor, and 28 mutations shared by most of the metastatic regions. The detection of private mutations suggested ongoing regional clonal evolution.
We inferred ancestral relationships and constructed a phylogenetic tree of the tumor regions by clonal ordering, as described by Merlo et al.20 (Fig. 2C), which revealed branching rather than linear tumor evolution. One branch evolved into the clones present in metastatic sites, and the other diversified into primary tumor regions. R4 shared some, but not all, primary-tumor and metastatic mutations, which suggested the presence of at least two clonal populations in this region that arose from progenitor cells of the metastases and of other primary tumor sites. Variant frequencies in the R4 ultradeep-sequencing data revealed that mutations shared with metastatic sites were detected at higher frequencies than were mutations shared with other primary-tumor regions, further supporting the presence of two subclones in R4 (Fig. 2 in the Supplementary Appendix). (For an exploratory phylogenetic analysis of the synonymous mutations, see Fig. 3 in the Supplementary Appendix.)
A single biopsy revealed an average of 70 somatic mutations, approximately 55% of all mutations detected in this tumor. Only 34% of all mutations that were detected by multiregion sequencing in the nephrectomy specimen were present in all regions (31% if pretreatment and metastasectomy samples were included), indicating that a single biopsy was not representative of the mutational landscape of the entire tumor bulk.
To address whether everolimus exposure may contribute to intratumor heterogeneity, we compared the phylogenetic relationships of pretreatment samples with those obtained after treatment samples (Fig. 2C). Of 71 mutations in pretreatment samples of the primary tumor, 67 were also present in post-treatment primary-tumor regions, and 64 of 66 mutations in the chest-wall metastases were present in post-treatment metastatic regions, indicating that the two main branches of the phylogenetic tree were present before drug treatment. Consistent with the post-treatment analysis, 60% of the mutations in pretreatment samples of the primary tumor and chest-wall metastases were not shared by both biopsy samples. Clones in R4 are unlikely to have evolved from pretreatment samples of the primary tumor or chest-wall metastases during therapy, since such evolution would have required the reversion of a large number of somatic mutations to wild-type, further supporting the presence of intratumor heterogeneity before treatment. Finally, samples taken before and after 6 and 12 weeks of everolimus exposure had similar numbers of nonsynonymous mutations (Fig. 4 in the Supplementary Appendix). Thus, everolimus does not appear to increase the mutational load, and the main phylogenetic branches were present in the tumor before treatment, indicating that intratumor heterogeneity was not a consequence of everolimus treatment.
Regional Ploidy Profiling and Chromosomal Aberration Detection
Ploidy profiling21 revealed a diploid profile for the majority of primary regions, whereas region m2b of the excised chest-wall metastasis harbored two subtetraploid populations (Fig. 2D). R4, the region most resembling the metastatic sites through clonal-ordering analysis, had a tetraploid profile, which suggests that the subtetraploid population in the chest-wall metastasis may have developed from a tetraploid intermediate in R4. Tumor regions were subjected to SNP-array–based allelicimbalance detection to identify chromosomal aberrations. Pretreatment samples of the primary tumor and metastasis were excluded because of insufficient DNA, and R1, R3, and R5 failed quality control. Sections of allelic imbalance on chromosome 3p were the only ubiquitous abnormalities (Fig. 5A in the Supplementary Appendix). Taken together with the corresponding reduced array signal intensities on chromosome 3p (Fig. 5B in the Supplementary Appendix), this indicated loss of heterozygosity through single deletion events in these 3p sections encoding VHL and the histone H3K36 methyltransferase SETD2. No tumor regions shared identical allelic-imbalance profiles, and heterogeneity of allelic imbalance within metastases, which is probably driven by aneuploidy, indicates that chromosomal aberrations contribute to genetic intratumor heterogeneity.
Intratumor Genetic Heterogeneity and Convergent Tumor Evolution
A comparison of genes that were mutated in this tumor with genes recurrently mutated in clear-cell carcinoma4,22 identified VHL, KDM5C, SETD2, and MTOR (Fig. 2B). Of these driver genes, only VHL was mutated ubiquitously in all analyzed regions. In contrast, SETD2 harbored three distinct mutations with different regional distributions (Fig. 2C): the metastases shared a missense mutation, R4 carried a splice-site mutation, and all other regions shared a 2-bp frameshift deletion, which was also detected in R4. Since SETD2 trimethylates H3K36, we stained several tumor regions with an antibody for trimethylated H3K36 to identify the consequences of mutational intratumor heterogeneity on protein function. Trimethylated H3K36 was reduced in cancer cells but positive in most stromal cells and in a SETD2 wild-type control clear-cell carcinoma (Fig. 6 in the Supplementary Appendix). These data support phenotypic convergent evolution through loss of SETD2 methyltransferase function driven by three distinct, regionally separated mutations on a background of ubiquitous loss of the other SETD2 allele on chromosome 3p.
Convergent evolution was observed for the X-chromosome–encoded histone H3K4 demethylase KDM5C, harboring disruptive mutations in R1 through R3, R5, and R8 through R9 (missense and frameshift deletion) and a splice-site mutation in the metastases (Fig. 2B and 2C).
mTOR Functional Intratumor Heterogeneity
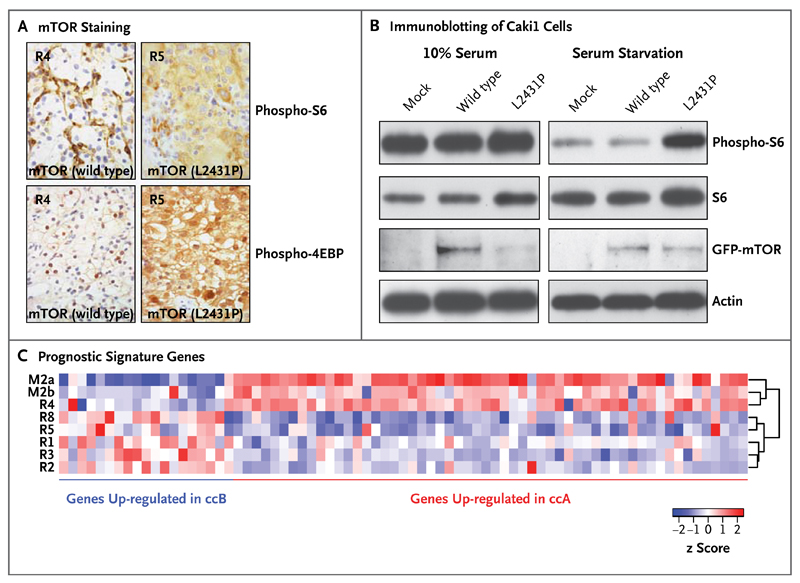
The mammalian target of rapamycin (mTOR) kinase carried a kinase-domain missense mutation (L2431P) in all primary tumor regions except R4. All tumor regions harboring mTOR (L2431P) had increased staining of the downstream mTOR pathway targets phospho-S6 and phospho-4EBP. Regions harboring wild-type mTOR had absent phospho-S6 and phospho-4EBP staining in tumor cells (Fig. 3A). It is unlikely that everolimus would affect activity in the mTOR pathway in these specimens, which were acquired 7 days after drug discontinuation (drug half-life, 30 hours).23 Transfection of complementary DNA encoding mTOR (L2431P) into a clear-cell carcinoma line enhanced phospho-S6 staining after serum starvation, indicating that L2431P promotes constitutive mTOR activation (Fig. 3B). Transient transfection of renal-cell carcinoma lines with the mutant mTOR construct did not affect everolimus sensitivity in vitro (data not shown). The mTOR sequence was aligned with the structurally related phosphatidylinositol 3 (PI3) kinase beta (Fig. 7 in the Supplementary Appendix). A structural model derived from this alignment suggests that L2431 maps to a hydrophobic pocket within an autoinhibitory domain adjacent to the activation loop. The substitution of L2431 by proline may affect the conformation of the mTOR activation loop. These data suggest that genetic intratumor heterogeneity is associated with functional heterogeneity of kinase activity.
Figure 3. Correlations between Genotype and Phenotype in Patient 1.
Panel A shows phospho-S6 (Ser235/236) and phospho-4EBP (Thr37/46) staining. All tumor regions harboring mTOR (L2431P) had increased staining of the downstream mTOR-pathway targets phospho-S6 and phospho-4EBP. Regions harboring wild-type mTOR had absent phospho-S6 and phospho-4EBP staining in tumor cells. Panel B shows immunoblotting of Caki1 cells (derived from a human renal-cell carcinoma) that were transiently transfected with green fluorescent protein (GFP) vector alone (mock), GFP-mTOR (wild type), or GFP-mTOR (L2431P) with and without serum starvation. Panel C shows hierarchical clustering of samples on the basis of prognostic signature genes of two molecular subgroups: clear-cell A (ccA), which indicates a good prognosis, and clear-cell B (ccB), which indicates a poor prognosis. The metastatic sites (M2a and M2b) and the primary-tumor site R4 segregated together, enriched for genes in the clear-cell A subgroup, in contrast to the remaining tumor regions that were enriched for the clear-cell B subgroup, showing that gene-expression signatures may not correctly predict outcomes if samples are obtained from a single biopsy. The brackets on the right side of the heat map (dendrogram) indicate the hierarchical clustering of the samples according to the expression of the analyzed genes. The z scores indicate the difference in standard deviations between the mRNA expression of a gene in a sample and its mean mRNA expression across all samples.
Intratumor Heterogeneity of a Prognostic Signature
We determined the intratumoral expression of a 110-gene signature shown to classify clear-cell carcinoma into two molecular subgroups: clear-cell A (associated with a good prognosis) and clear-cell B (associated with a poor prognosis).24 Consistent with the phylogenetic analysis, metastatic sites and the primary-tumor site R4 segregated together, enriched for genes in the clear-cell A subgroup, in contrast to the remaining tumor regions that were enriched for the clear-cell B subgroup (Fig. 3C). Thus, prognostic gene-expression signatures may not correctly predict outcomes if they are assessed from a single region of a heterogeneous tumor.
Intratumor Genetic Heterogeneity in Three Consecutive Tumors
To determine whether intratumor heterogeneity was present in consecutive clear-cell carcinomas from the E-PREDICT trial, we performed multiregion exome sequencing on the primary tumor and a metastasis from Patient 2 and ploidy and allelicimbalance profiling on primary tumors from Patients 2, 3, and 4. CT imaging showed no change in tumor dimensions during 6 weeks of everolimus treatment.
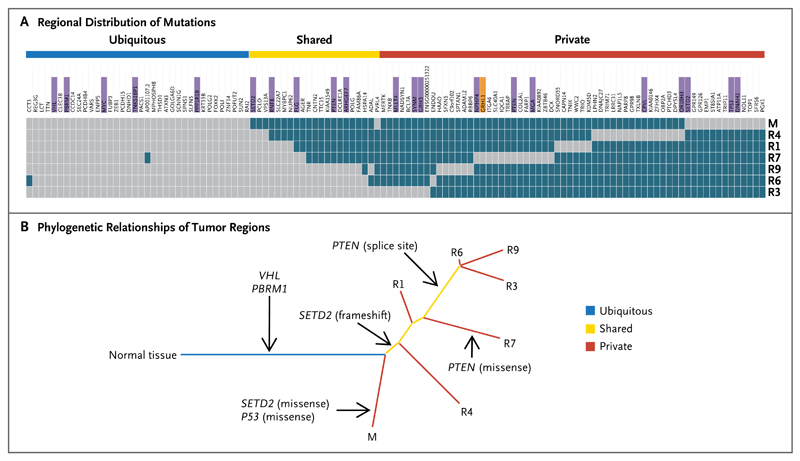
Patient 2 had a metastatic tumor with a 1-bp deletion in VHL. Pretreatment core biopsy samples contained less than 5% tumor cells and were excluded. Primary-tumor regions from R1 through R9 were harvested from the nephrectomy specimen. After 5 months of participation in the trial with no objective tumor response, a core biopsy specimen was obtained from a progressive liver metastasis. We performed exon-capture sequencing on tumor and germline DNA samples (median coverage, 61 reads) (Table 1 in the Supplementary Appendix). R2, R5, and R8 were excluded from the analysis because of low variant-to-total-read ratios, indicating high stromal contamination. A total of 119 somatic mutations were detected and their regional distribution mapped (Fig. 4A, and Table 4 in the Supplementary Appendix). We used Sanger sequencing to validate somatic mutations in the tumor-suppressor genes VHL, PBRM1, and TP53, in 2 independent and spatially separated mutations in SETD2 (missense and frameshift mutations) and PTEN (splice-site and missense mutations), and in 14 of 15 randomly selected mutations (validation rate, 95%) (Fig. 8 in the Supplementary Appendix).
Somatic Mutational Heterogeneity
Phylogenetic analysis of tumor from Patient 2 revealed a branching pattern (Fig. 4B), which was consistent with the findings from primary tumor and metastases obtained from Patient 1. Only 37% of the somatic mutations that were identified in the nephrectomy specimen were ubiquitously detectable in all regions (31% if the metastasis, which was biopsied at the time of progression, was included). Prolonged everolimus exposure was not associated with an increase in the number of nonsynonymous mutations in the liver metastasis, as compared with the nephrectomy-biopsy specimens (Fig. 9 in the Supplementary Appendix), further indicating that everolimus does not increase the mutational load.
Figure 4. Genetic Intratumor Heterogeneity and Phylogeny in Patient 2.
Panel A shows the regional distribution of somatic mutations detected by exome sequencing in a heat map, with gray indicating the presence of a mutation and dark blue the absence of a mutation. The color bars above the heat map indicate classification of mutations according to whether they are ubiquitous, shared by primary-tumor regions, or unique to the region (private). For gene names, purple indicates that the mutation was validated, and orange indicates that the validation of the mutation failed. Panel B shows phylogenetic relationships of the tumor regions. Branch lengths are proportional to the number of somatic mutations separating the branching points. Potential driver mutations were acquired by the indicated genes in the branch (arrows).
There was no evidence of intratumor heterogeneity on ploidy profiling in primary tumor from Patient 2 (Fig. 10 in the Supplementary Appendix). However, allelic-imbalance analysis (with the exception of R8 and liver metastasis, which were excluded because of insufficient DNA) identified multiple heterogeneous chromosomal aberrations suggestive of intratumor heterogeneity. Allelic imbalances on chromosomes 3p and 10q were the only ubiquitous aberrations (Fig. 11 in the Supplementary Appendix).
Trimethylated H3K36 staining was absent from tumor cells in regions with SETD2 frameshift or missense mutations (Fig. 12 in the Supplementary Appendix), indicating that both mutations together with a 3p deletion confer convergent loss of function. Regions with either a splice-site mutation or a missense mutation in PTEN, a negative regulator of the PI3 kinase–Akt pathway located on chromosome 10, showed increased phospho-Akt staining, as compared with PTEN wild-type regions (Fig. 13 in the Supplementary Appendix), consistent with loss of PTEN function and convergent phenotypic evolution.
Regional allelic-imbalance profiling of primary tumors from Patients 3 and 4 provided further evidence of genetic intratumor heterogeneity (Fig. 11 in the Supplementary Appendix). Only 4 of 30 samples from four patients had identical allelicimbalance profiles (tumor from Patient 3 in R1, R3, R4, and R9). Chromosome 3p aberrations occurred ubiquitously in all regions from all tumors, and allelic-imbalances of 10q (in tumor from Patient 2) and in 5q and 6q (in tumor from Patient 4) were ubiquitously present in one case each. These early ubiquitous events were outnumbered by nonubiquitous aberrations, indicating that the majority of chromosomal events occurred after tumors diverged, providing further evidence of branching evolution. Ploidy profiling detected intratumor heterogeneity in tumor from Patient 4 (Fig. 10 in the Supplementary Appendix), and Sanger sequencing of SETD2 in Patients 3 and 4 revealed intratumor heterogeneity in Patient 4: seven regions of tumor sharing a SETD2 frameshift mutation harbored absent trimethylated H3K36 staining, whereas a single region with wild-type SETD2 but mutant VHL harbored strong tumor-cell trimethylated H3K36 staining (Fig. 14 in the Supplementary Appendix).
Discussion
Multiregion genetic analysis of four consecutive tumors provided evidence of intratumor heterogeneity in every tumor, with spatially separated heterogeneous somatic mutations and chromosomal imbalances leading to phenotypic intratumor diversity (activating mutation in MTOR) and uniformity (loss-of-function mutation in SETD2 and PTEN). Of all somatic mutations found on multiregion sequencing, 63 to 69% were heterogeneous and thus not detectable in every sequenced region. Heterogeneous patterns of allelic imbalance were found in all tumors, and ploidy heterogeneity was found in two tumors. Therefore, we found that a single tumor-biopsy specimen reveals a minority of genetic aberrations (including mutations, allelic imbalance, and ploidy) that are present in an entire tumor.
Pretreatment tumor-biopsy specimens from Patient 1 had branched mutational profiles that were almost identical to those detected after everolimus exposure. Everolimus is not known to be mutagenic, and the number of nonsynonymous mutations did not increase after the administration of everolimus. Intratumor heterogeneity is unlikely to be confounded by clonal selection, since no tumor responses were observed during the brief preoperative treatment period. Since intratumor heterogeneity preceded therapy, it is unlikely that treatment biased the interpretation of these analyses.
An unexpected finding was the detection of spatially separated distinct somatic mutations affecting the histone H3K36 methyltransferase SETD2, the histone H3K4 demethylase KDM5C, and the negative regulator of the PI3 kinase–Akt pathway, PTEN. Despite genetic divergence during tumor progression, phenotypic convergent evolution occurs, indicating a high degree of mutational diversity, a substrate for Darwinian selection, and evolutionary adaptation.
The heterogeneous MTOR mutation renders the kinase constitutively active, increasing S6 phosphorylation, a potential biomarker of response to mTOR inhibitors in clear-cell carcinoma.25 Such spatially separated somatic mutations altering pathway activity suggest that multiregional analyses may be required to predict the therapeutic outcome. Further studies will assess whether activating mutations in MTOR depend on this pathway, which may result in higher responsiveness to mTOR inhibition.
Intratumor heterogeneity was evident at the RNA-expression level, with expression signatures of good or poor prognosis detected in different regions of the same tumor. Although the 7-day washout period minimized a direct influence of everolimus on prognostic signature expression, we cannot exclude the possibility that potential unknown and persistent effects of everolimus pretreatment on the tumor–stroma composition might alter mRNA expression. The prognostic signature was described to be independent of metastatic stage,24 consistent with the occurrence of the good prognostic signature in metastases.
Branched tumor evolution underscores the importance of targeting ubiquitous alterations in the trunk of the phylogenetic tree. Such ubiquitous allelic-imbalance events were seen on chromosome 3p (encoding VHL, PBRM1, and SETD2), 5q, 6q, and 10q. Larger multiregional series will probably identify genes that can be targeted in the trunks of the phylogenetic tree for clear-cell carcinoma. Intratumor heterogeneity within the primary tumor may account for the benefits associated with cytoreductive nephrectomy26–28 by eliminating an evolutionary reservoir of phenotypic tumor-cell diversity.
Genomics analyses from single tumor-biopsy specimens may underestimate the mutational burden of heterogeneous tumors. Intratumor heterogeneity may explain the difficulties encountered in the validation of oncology biomarkers owing to sampling bias,29 contribute to Darwinian selection of preexisting drug-resistant clones,12,30 and predict therapeutic resistance.13 Reconstructing tumor clonal architectures and the identification of common mutations located in the trunk of the phylogenetic tree may contribute to more robust biomarkers and therapeutic approaches.
Supplementary Material
Acknowledgments
Supported by grants from the Medical Research Council, Cancer Research UK, the Royal Marsden Hospital Renal Research Fund, Novartis, EU Framework 7 Personalized RNA Interference to Enhance the Delivery of Individualized Cytotoxic and Targeted Therapeutics (PREDICT), and the Wellcome Trust (to Dr. Futreal).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
We thank the patients; Nasir Khan, Isobelle Coombes, Kim Edmonds, Amy Thomas, Jade Griffiths, Phil Clarke, Maggie James, and Peter Campbell; and the U.K. National Institute for Health Research.
This article is dedicated to Tim Christmas.
References
- 1.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sjöblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 4.Varela I, Tarpey P, Raine K, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–42. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–66. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell PJ, Yachida S, Mudie LJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–13. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell PJ, Pleasance ED, Stephens PJ, et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci U S A. 2008;105:13081–6. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mullighan CG, Phillips LA, Su X, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–80. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roche-Lestienne C, Soenen-Cornu V, Grardel-Duflos N, et al. Several types of mutations of the Abl gene can be found in chronic myeloid leukemia patients resistant to STI571, and they can pre-exist to the onset of treatment. Blood. 2002;100:1014–8. doi: 10.1182/blood.v100.3.1014. [DOI] [PubMed] [Google Scholar]
- 11.Shah NP, Nicoll JM, Nagar B, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–25. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 12.Inukai M, Toyooka S, Ito S, et al. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non-small cell lung cancer. Cancer Res. 2006;66:7854–8. doi: 10.1158/0008-5472.CAN-06-1951. [DOI] [PubMed] [Google Scholar]
- 13.Lee AJ, Endesfelder D, Rowan AJ, et al. Chromosomal instability confers intrinsic multi-drug resistance. Cancer Res. 2011;71:1858–70. doi: 10.1158/0008-5472.CAN-10-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerlinger M, Swanton C. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br J Cancer. 2010;103:1139–43. doi: 10.1038/sj.bjc.6605912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Navin N, Kendall J, Troge J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–4. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding L, Ellis MJ, Li S, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yachida S, Jones S, Bozic I, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tao Y, Ruan J, Yeh SH, et al. Rapid growth of a hepatocellular carcinoma and the driving mutations revealed by cell-population genetic analysis of whole-genome data. Proc Natl Acad Sci U S A. 2011;108:12042–7. doi: 10.1073/pnas.1108715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gnirke A, Melnikov A, Maguire J, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–9. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–35. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 21.Navin N, Krasnitz A, Rodgers L, et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010;20:68–80. doi: 10.1101/gr.099622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalgliesh GL, Furge K, Greenman C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–3. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Donnell A, Faivre S, Burris HA, III, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral mammalian target of rapamycin inhibitor everolimus in patients with advanced solid tumors. J Clin Oncol. 2008;26:1588–95. doi: 10.1200/JCO.2007.14.0988. [DOI] [PubMed] [Google Scholar]
- 24.Brannon AR, Reddy A, Seiler M, et al. Molecular stratification of clear cell renal cell carcinoma by consensus clustering reveals distinct subtypes and survival patterns. Genes Cancer. 2010;1:152–63. doi: 10.1177/1947601909359929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho D, Signoretti S, Dabora S, et al. Potential histologic and molecular predictors of response to temsirolimus in patients with advanced renal cell carcinoma. Clin Genitourin Cancer. 2007;5:379–85. doi: 10.3816/CGC.2007.n.020. [DOI] [PubMed] [Google Scholar]
- 26.Flanigan RC, Mickisch G, Sylvester R, Tangen C, Van Poppel H, Crawford ED. Cytoreductive nephrectomy in patients with metastatic renal cancer: a combined analysis. J Urol. 2004;171:1071–6. doi: 10.1097/01.ju.0000110610.61545.ae. [DOI] [PubMed] [Google Scholar]
- 27.Flanigan RC, Salmon SE, Blumenstein BA, et al. Nephrectomy followed by interferon alfa-2b compared with interferon alfa-2b alone for metastatic renal-cell cancer. N Engl J Med. 2001;345:1655–9. doi: 10.1056/NEJMoa003013. [DOI] [PubMed] [Google Scholar]
- 28.Mickisch GH, Garin A, van Poppel H, de Prijck L, Sylvester R. Radical nephrectomy plus interferon-alfa-based immunotherapy compared with interferon alfa alone in metastatic renal-cell carcinoma: a randomised trial. Lancet. 2001;358:966–70. doi: 10.1016/s0140-6736(01)06103-7. [DOI] [PubMed] [Google Scholar]
- 29.Poste G. Bring on the biomarkers. Nature. 2011;469:156–7. doi: 10.1038/469156a. [DOI] [PubMed] [Google Scholar]
- 30.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–77. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.