Figure 1. Sequence alignment and genetic organization of the periplasmic copper chaperone PccA in R. capsulatus.
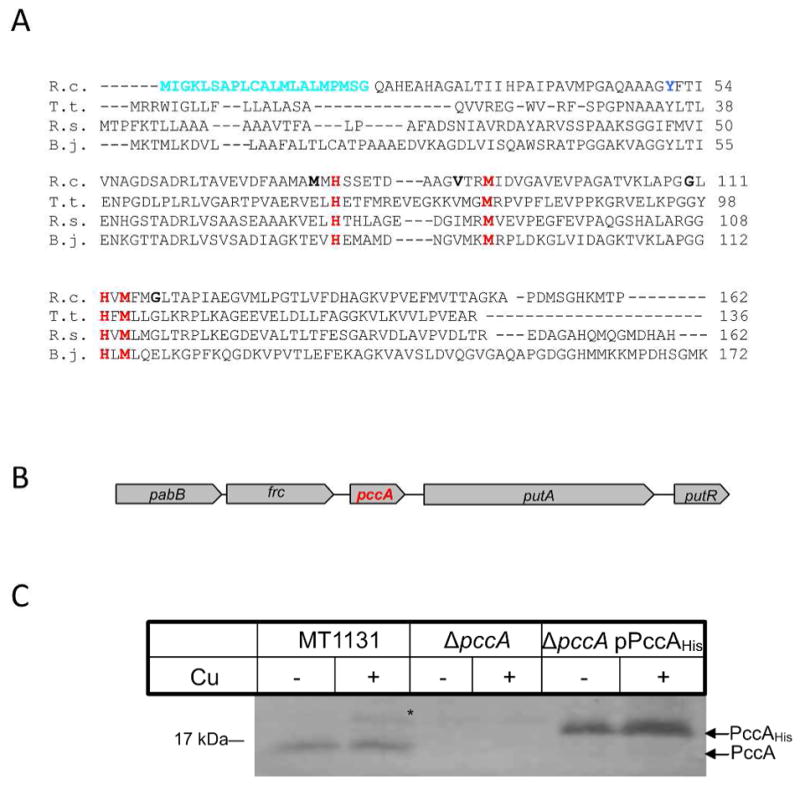
(A) The predicted amino acid sequence of PccA of R. capsulatus (R.c.) was aligned to other members of the PCuAC copper chaperone family using the Clustal sequence alignment algorithm: T. thermophilus PCuAC (T.t.); R. sphaeroides RSP_2017 (R.s.); and B. japonicum PcuC (B.j.). Indicated are the predicted cleavable signal sequence of R. capsulatus PccA (cyan) and the conserved residues of the putative Cu-binding site (red). The single tyrosine residue used for measuring the intrincisc fluorescence of PccA is indicated in blue and the positions used for cysteine substitutions in PccA are indicated in bold. (B) Genetic organization of pccA in R. capsulatus. The nucleotide sequence of R. capsulatus was retrived from NCBI and analyzed using the NCBI tool box (www.ncbi.nlm.nih.gov). Displayed is the R. capsulatus genomic region encompassing pabB, p-amino-benzoat synthase (Rcc02644); frc, CoA transferase (Rcc02645); pccA (Rcc02646) putA, proline dehydrogenase (Rcc02647) and putR, proline dehydrogenase regulator (Rcc02648). (C) Wild type R. capsulatus cells (MT1131), cells of the ΔpccA strain and of the ΔpccA strain carrying pccA on a low copy plasmid (ΔpccA/pPccAHis) were grown on MPYE medium or on MPYE medium supplemented with 10 μM CuSO4. Cells were harvested, and after washing, TCA precipitated and separated on SDS-PAGE. After western blotting, the samples were decorated with α-PccA antibodies. *indicates a band that cross-reacted with the α-PccA antibody in wild type cells grown in the presence of Cu.
