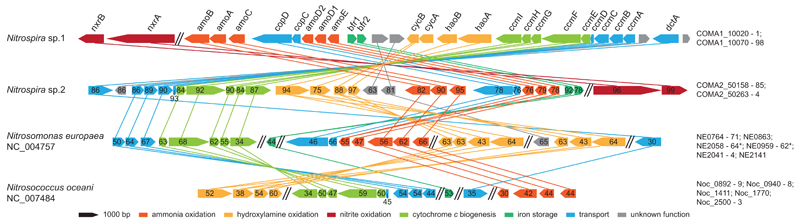
Figure 3. Schematic illustration of the AMO genomic region in Nitrospira and selected AOB.
The AMO locus in Nitrospira sp.1 in comparison to sp.2 and the beta- and gammaproteobacterial AOB Nitrosomonas europaea and Nitrosococcus oceani, respectively. The position of NXR on the AMO-containing Nitrospira contigs is also indicated. Homologous genes are connected by lines. Functions of the encoded proteins are represented by colour, the arrow shows direction of transcription. Numbers specify amino acid identities to Nitrospira sp.1. Parallel double lines designate a break in locus organization. Locus tags for each organism are listed on the right. Genes are drawn to scale. amo, ammonia monooxygenase; bfr; bacterioferritin; ccm, cytochrome c biogenesis; cop, copper transport; cyc, cytochrome c; dct, sodium:dicarboxylate symporter; hao, hydroxylamine dehydrogenase; nxr, nitrite oxidoreductase.