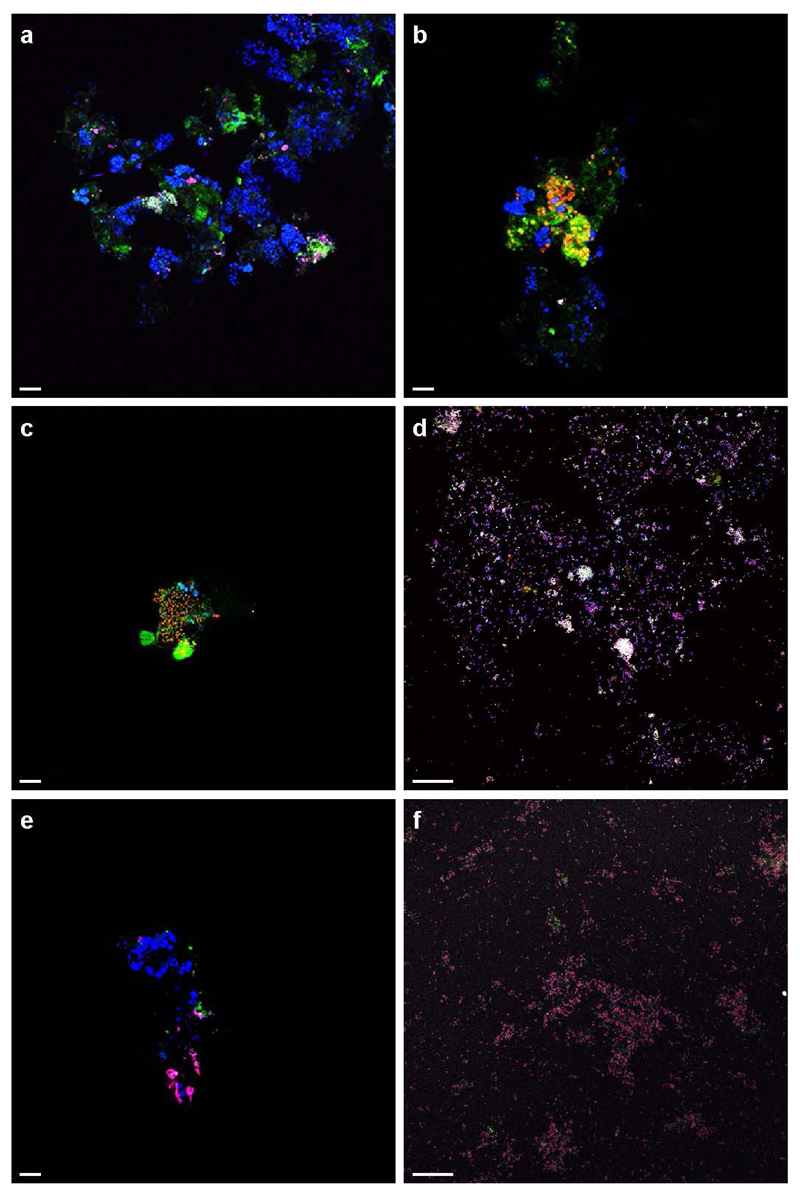
Extended Data Figure 5. Control experiments of AMO-labelling.
a, Cells incubated with the fluorescent dye FTCP (green) were stained by FISH using probes specific for Nitrospira (Ntspa662, red) and all bacteria (EUB338mix, blue). A small cell cluster was stained by FTCP and targeted by both probes (resulting in a white overlay signal), while all other bacteria (in blue) were not or only slightly stained by FTCP. The green signal is due to autofluorescence and unspecific FTCP binding to the floc matrix. b, Anammox cells (Amx820, blue) showed minor staining by FTCP (green), but to a much lesser degree than Nitrospira (Ntspa662, red; yellow overlay). c and d, Positive controls: (c) ammonium oxidizing bacteria (Nso1225 and Nso190, red) in an aerobic enrichment culture and (d) a Nitrosomonas europaea pure culture (NEU, red, and EUB338mix, blue) were stained by FTCP (resulting in yellow and white overlays, respectively). e and f, Negative controls: (e) canonical Nitrospira in an aerobic enrichment culture (Ntspa662, blue) and (f) a Nitrospira moscoviensis pure culture (Ntspa662, red, and EUB338mix, blue; magenta overlay) did not show any labelling with FTCP (green). The two bright green structures in (c) and the bright pink signal in (e) are due to autofluorescence. Images are representative of two (a and b) or one (c to f) individual experiments, with three technical replicates each. Scale bars in all panels represent 10 μm.