Figure 1.
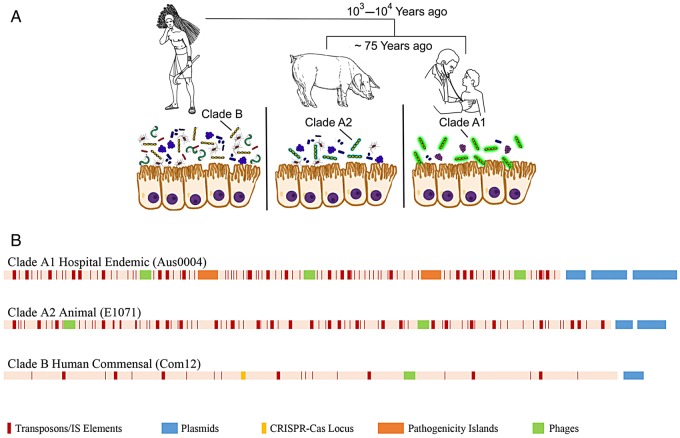
Human impact on the evolution of Enterococcus faecium. A, The species E. faecium split into 2 distinct species in parallel with the urbanization of humans and the spread of animal domestication, and then the animal lineage split again coincident with the application of antibiotics in agriculture and human medicine. Human commensal strains evolved to exist in the complex native consortium of the human gastrointestinal tract. Animal strains largely derive from domesticated animals, to which the majority of antibiotics produced in the United States are applied at low levels for most of the life of the animal, with enterococcal strains adapting to fill opportunities in simplified consortia [20]. Hospital-endemic strains are well adapted to life in the simplified antibiotic-treated patient consortium and to survival outside of the patient in hospitals. B, Linear depiction of representative genomes of E. faecium from clades A1 (Aus004 [16]), A2 (E1071 [6]), and B (Com12 [21]), showing the impact of human intervention on their rapid evolution, which highlights the accretion of mobile genetic elements in clades A1 and A2 that are rare in human commensal strains largely possessing CRISPR/CAS loci.