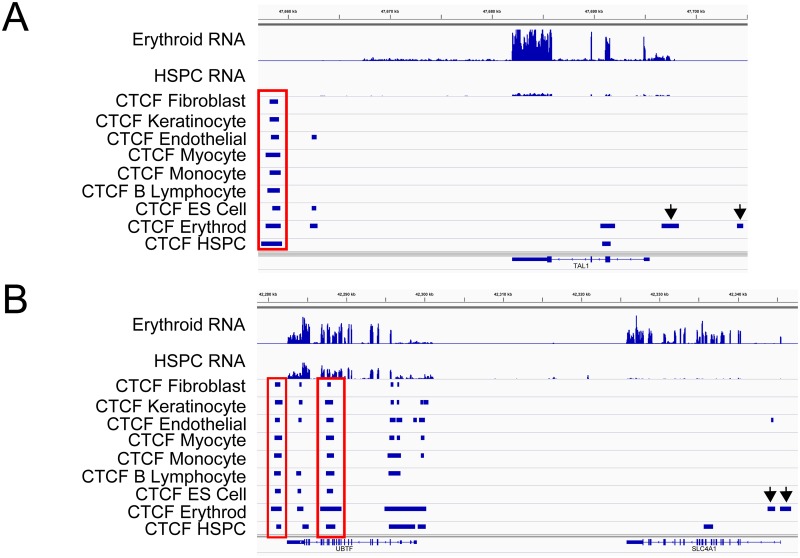
Fig 2. Invariant and cell type-specific CTFC sites.
Patterns of CTCF occupancy in HSPC and erythroid cell chromatin were compared to CTCF occupancy in several human ENCODE ChIP-seq data sets, including fibroblast, keratinocyte, endothelial, myocyte, monocyte, lymphocyte, embryonic stem (ES) cell, erythroid and HSPC cells. A. At the TAL1 locus, a 3’ site of invariant CTCF binding marked by the rectangle is present in all cell types. Two sites of erythroid-specific CTCF binding, denoted by the arrows, are present 5’ of the gene. Corresponding RNA-seq tracks in HSPC and erythroid cells are shown at the top. Genomic coordinates: Chr1:47,600–47,700. B. At the UBTF and SLC4A1 loci, there are 2 sets of invariant CTCF binding, marked by rectangles, 3’ of the UBTF locus, present in all cell types. One site of erythroid-specific CTCF binding, denoted by the arrows, are present 5’ of the SLC4A1 locus. Corresponding RNA-seq tracks in HSPC and erythroid cells are shown at the top. Genomic coordinates: Chr17:42,280–42,340.