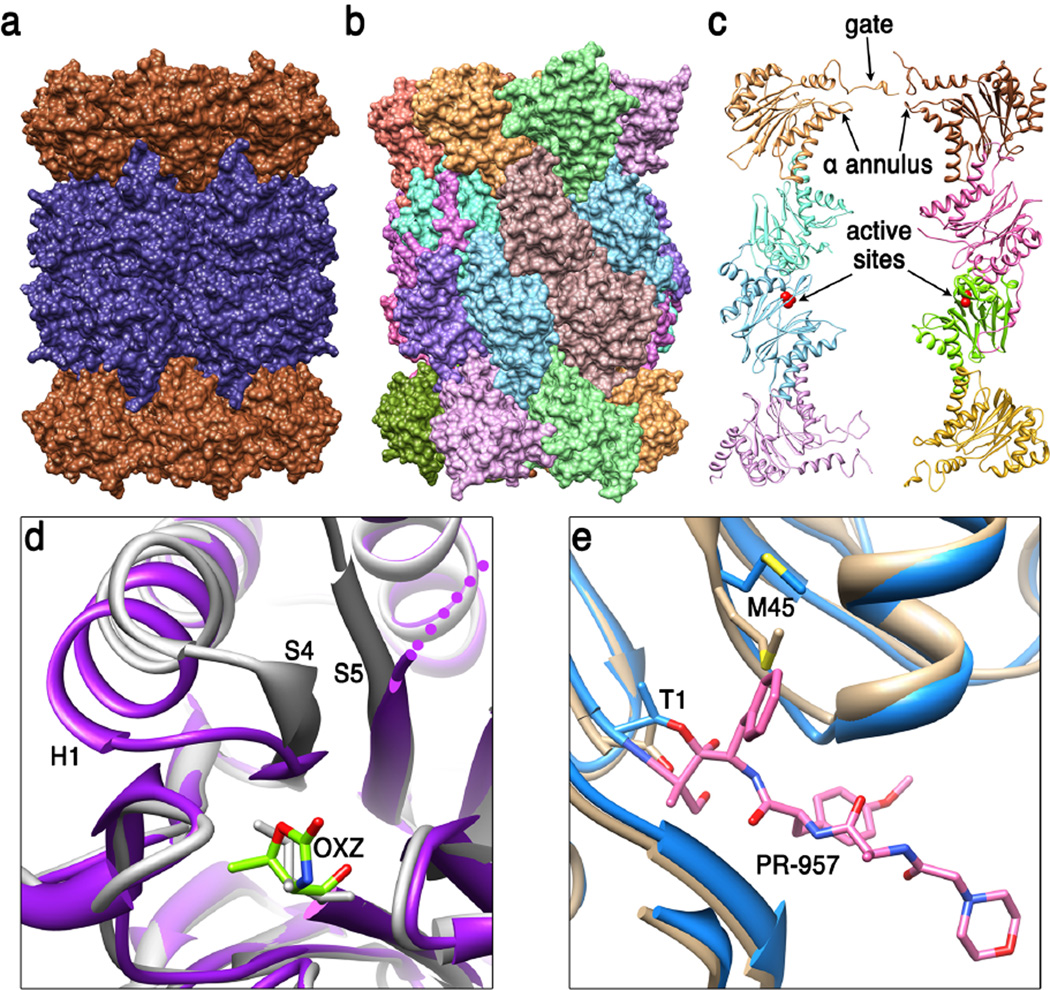
Figure 1. 20S proteasome core particle (20S CP).
(a) Side view of the archaeal T. acidophilum 20S CP (51) (pdb code 1pma). End rings comprise seven identical α subunits (brown) and the two middle rings comprise seven identical β subunits (dark blue). (b) Side view of the eukaryotic S. cerevisiae 20S CP (24) (pdb code 1ryp). Each of the seven different α subunits and seven different β subunits occupies a unique position within their respective rings. The whole structure has two-fold symmetry relating the top and bottom halves of the structure to each other, with the 2-fold axis in the horizontal plane, a little to the right of center in this view. (c) Cutaway view showing internal features. The S. cerevisiae 20S CP is shown in ribbon representation with just eight subunits displayed in order to reveal the hollow interior. Labeled features include residues that contribute to the asymmetric closed gate structure, loops that contribute to the α annulus, and the active sites of β1 and β5 in the lower β ring (only the β1, β2, and β5 subunits have active sites in eukaryotic proteasomes). (d) Conformational changes at the activate site of M. tuberculosis 20S CP that are induced upon binding of inhibitor suggest the possibility of developing a specific therapeutic (49). The loop connecting S4 and H1 of the β subunit moves from the unbound conformation (white, pdb code 2fhg) to cover OXZ, the inhibitor oxazolidin-2-one ring on Thr1 in the stabilized complex (purple, pdb code 3h6f). (e) Comparison of mouse liver constitutive and inducible β5 S1 binding pocket (30). Met45 adopts the sky blue conformation (pdb code 3unf) when bound to the PR-957 inhibitor (pink), which binds with a large hydrophobic group in the S1 pocket. Met45 also adopts this conformation in the unbound immunoproteasome but adopts the tan conformation in the unbound constitutive proteasome (pdb code 3une). This requirement for repositioning Met45 explains why immunoproteasomes prefer to cleave substrate after large hydrophobic side chains.