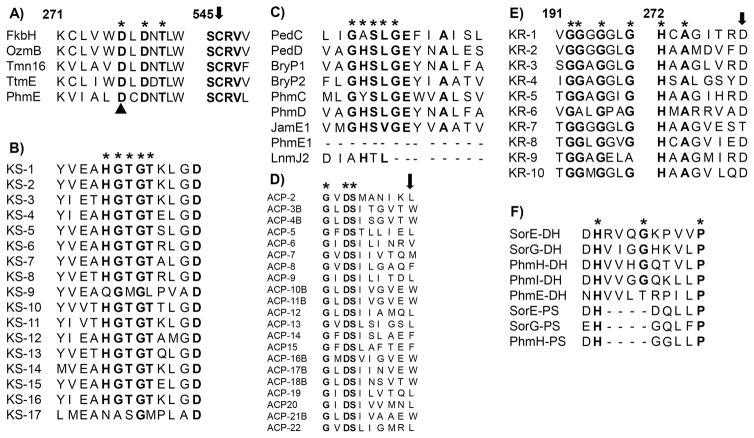
Figure 3.
Sequence alignments of conserved domains of the deduced amino acid sequence of genes in the phormidolide gene cluster. A) Domain alignment of FkbH-like phosphatases. PhmE FkbH domain contains the DXDX(T/V) consensus sequence * and catalytic aspartate (black triangle) and cysteine (black arrow) proposed to dephosphorylate and attach the phosphoglyceryl starter unit. Numbering based on the Tmn16 (AB193609) sequence. OzmB, ABA39082; TtmE, DQ088168; FkbH, AAF86387. B) Domain alignment of 17 KS domains from Phm pathway. The conserved sequence with catalytic His HGTGT is denoted by *, which is absent from predicted non-elongating KS0 (KS-9, KS-17). C) AT domain alignment of trans-ATs from the pederin (PedC, AAS47559; PedD, AAS47563) and bryostatin (BryP, ABM63531) pathways, a cis-AT from the jamaicamide A pathway (JamE, AAS98777) and a predicted AT-docking domain from the leinamycin (Lnm, AAN85523) pathway. Predicted trans-ATs PhmC and PhmD align well with trans- and cis-ATs, while the predicted AT-docking domain (PhmE-AT1) is truncated. The malonate-specific motif GHS[LVIFAM]G is denoted by *. D) ACP domain alignment. The active site sequence GXDS is denoted by * and the conserved tryptophan in ACPs associated with β-branching is identified with a black arrow. A “B” denotes ACPs predicted to be involved in β-branched methylation. E) KR domain alignment, including the conserved D1758 in B-type KRs. The conserved NADPH binding motif CGGXGXXG is denoted by *. The conserved motif HXAXXXXD is denoted by * and a black arrow indicates an amino acid motif of B-type KRs that generates the D-3-hydroxyacyl moiety. F) Phm DH and PS domains aligned to DH and PS domains from sorangicin (SorE, ADN68480; SorG, ADN68482). The conserved DH active-site motif HXXXGXXXXP is denoted by *.