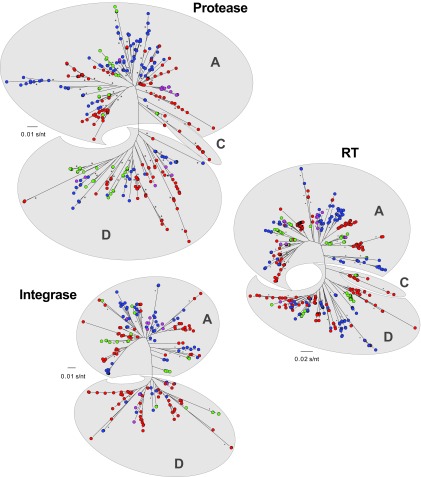
FIG 5.
Neighbor-joining phylogenetic trees were constructed using reads with a frequency of ≥10 corresponding to 105-bp fragments from the protease, RT, and integrase regions. Each color-coded dot represents a unique variant (frequency not depicted) in each patient corresponding to groups I (blue), II (red), III (green), and IV (purple). HIV-1 subtype-specific clusters are depicted by clouds labeled for each subtype, i.e., A, C, or D. Bootstrap resampling (1,000 data sets) of the multiple alignments tested the statistical robustness of the trees, with percentages above 75% indicated by asterisks. s/nt, substitutions per nucleotide.