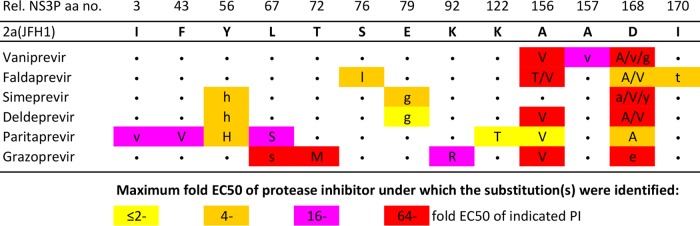
FIG 2.
Substitutions identified in the NS3Ps of HCV genotype 2a escape viruses during treatment with newer PIs. Huh7.5 cells were infected with the genotype 2a (isolate JFH1) virus, treated with vaniprevir, faldaprevir, simeprevir, deldeprevir, paritaprevir, or grazoprevir, and followed as described in Materials and Methods and in Fig. S3 in the supplemental material. At the time of viral escape, the NS3Ps of viruses were amplified using reverse transcription-PCR and were directly sequenced. Only NS3P positions at which a substitution was identified for at least one of the PIs are shown. Rel. NS3P aa no., NS3 protease amino acid number relative to that in the genotype 1a reference strain H77 (GenBank accession no. AF009606). The original genotype 2a (isolate JFH1) virus amino acid residues are used as references in this alignment. For each of the PIs studied, a dot indicates that the amino acid residue at the respective position did not change. Substitutions occurring under PI treatment are indicated by colored rectangles. The color of the rectangle indicates the highest multiple of the EC50 of the PI at which the substitution was identified. The letter(s) in each colored rectangle indicates the substitution(s) identified in escape variants, and different substitutions identified at the same position are separated by a slash. The substitutions estimated to be present in at least 50% of viral genomes are indicated by capital letters, while those estimated to be present in a minor percentage of viral genomes are indicated by lowercase letters. Substitutions that were also identified in nontreated cultures are not shown. For further details, see Tables S18 to S23 in the supplemental material.