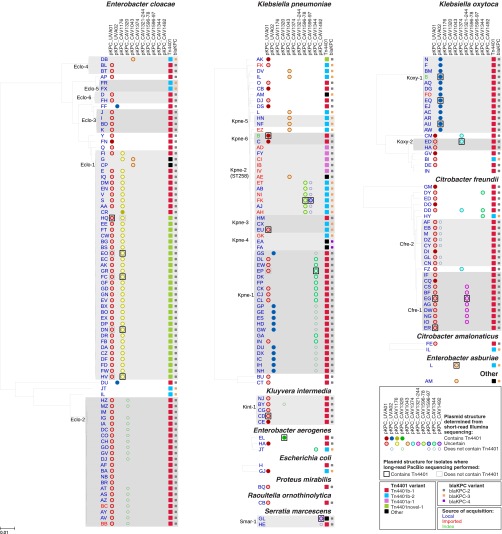
FIG 1.
Diversity of bacterial species, strains, plasmids, and Tn4401 variants. For each species, a phylogeny was generated from mapping to a species-specific chromosomal reference, after the deduplication of closely related isolates from the same patient (see Materials and Methods). Distinct strains are defined by a cutoff of ∼500 SNVs (see Materials and Methods); strains found in more than one patient are shaded gray. Circles show plasmid “presence” as determined from Illumina data, with the fill color indicating uncertainty about whether the plasmid contains blaKPC. Boxes show plasmid structures determined from long-read PacBio sequencing of 17 randomly chosen isolates, as well as the previously sequenced isolates from index patient B (37). Where the PacBio-sequenced isolate was excluded from the phylogeny as a patient duplicate, the plasmid structure of the corresponding closely related isolate from the same patient is shown. Tn4401 and blaKPC variants (Table 2) are indicated by large and small squares, respectively. The likely sources of blaKPC acquisition, as determined from epidemiological data, are indicated by text color.