FIG 5.
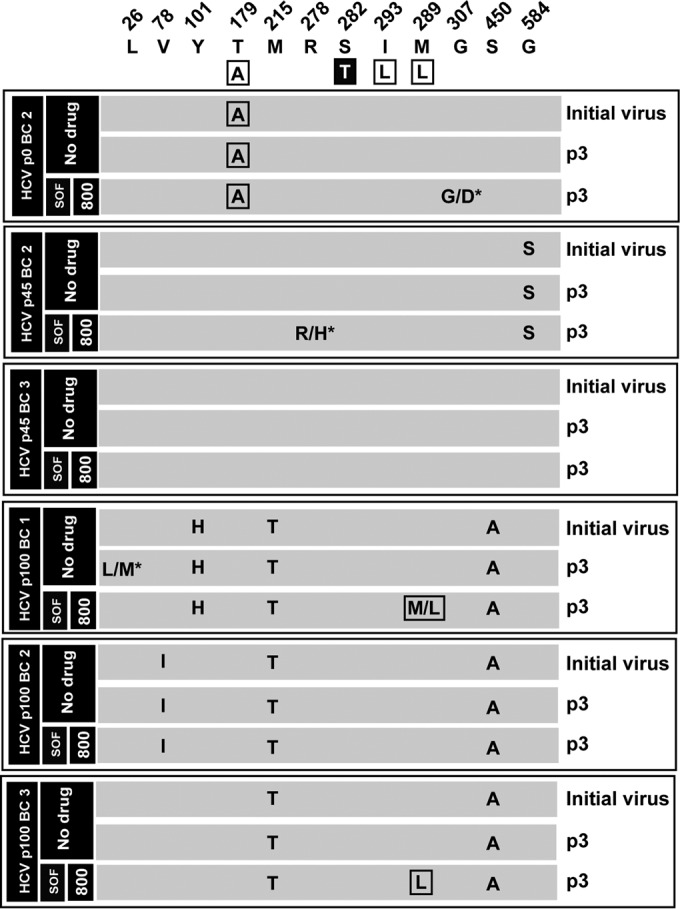
Amino acid substitutions in the consensus sequence of the NS5B (polymerase)-coding region of biological clones of hepatitis C virus passaged in the absence or presence of sofosbuvir. The parental viral populations from which the corresponding biological clones were retrieved (identified with the same code used in Fig. 4) and the sofosbuvir (SOF) concentration (nanomolar) are indicated in the filled boxes on the left; the virus passage number (p) is given in the last column. Biological clones not included in the nucleotide sequence analysis are those for which no sufficient viral RNA was obtained (compare with Fig. 4). The upper row includes the amino acids where substitutions have been found and their positions in the NS5B protein. The boxed amino acids in the second row are those that have been related to SOF resistance in HCV genotype 2a, with the major resistance substitution S282T highlighted. The six panels below the sequence describe the amino acid substitutions in each population, with those that have been related to SOF resistance boxed. A slash between two unmarked amino acids means that both were present at about 50% frequency in the consensus sequence according to the peaks in the sequencing data; a slash with an asterisk indicates dominance (around 70% frequency) of the first amino acid in the pair. The complete repertoire of synonymous and nonsynonymous mutations found in the viral populations is listed in Table S2 in the supplemental material. Procedures for nucleotide sequencing are described in Materials and Methods.
