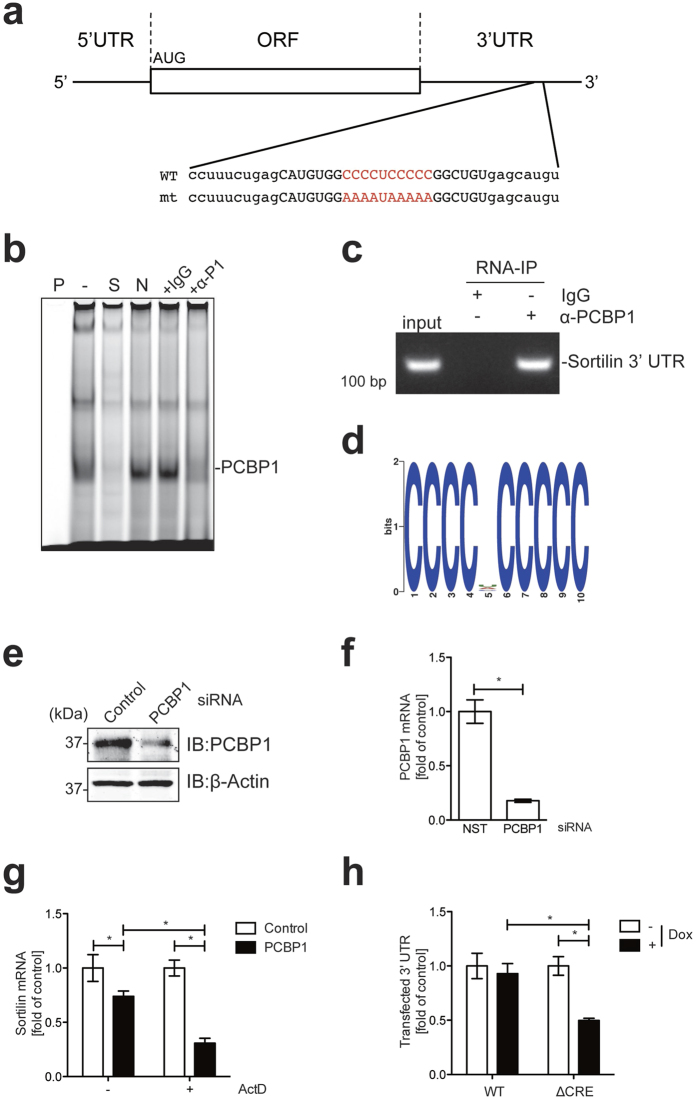
Figure 5. PCBP1 binds to C-rich elements in the 3′UTR of sortilin mRNA.
(a) Scheme of mouse sortilin mRNA containing defined mutations of the PCBP1-binding site. Only the sequence around the CRE in the 3′ UTR is shown. Synthetic RNA probes for RNA EMSA are shown as upper-case letters and CREs are in red. (b) RNA EMSA analysis. An IR800-conjugated WT CRE probe was mixed with binding buffer alone (P) or whole cell lysates from macrophages (−). Samples were separated by PAGE. Five-fold molar excess amounts of unlabeled WT CRE and mt CRE were added as specific competitor (S) and nonspecific competitor (N), respectively. Antibody supershift experiments were performed with anti-PCBP1 antibody (+α-P1). Rabbit IgG (+IgG) was used as a negative control in antibody supershift experiments. (c) RNA immunoprecipitation (RIP) analysis of PCBP1. RT-PCR was performed with RNA immunoprecipitated by anti-PCBP1 antibody to amplify the CRE-containing region of the 3′ UTR of sortilin mRNA. RNA immunoprecipitated by rabbit IgG (IgG) was used as a negative control. (d) Depiction of sequence significantly enriched by RIP-seq analysis as a position weight matrix (E-value = 7.8e-80). (e,f) Depletion of PCBP1 in RAW264.7 cells. Cells were transfected with siRNAs against PCBP1 or with a control non-targeting siRNA, and were harvested 2 d after transfection. Cell lysates were subjected to immunoblotting (e) with the indicated antibodies, and total RNAs were subjected to qRT-PCR (f) to quantify PCBP1 mRNA. Data are mean ± SD (n = 3). *p < 0.01. (g) Destabilization of sortilin mRNA by PCBP1 depletion in cells. RAW264.7 cells were transfected with control or PCBP1 siRNA for 48 h and then treated with 1 μg/ml ActD for 9 h. Total RNAs were subjected to qRT-PCR to quantify sortilin mRNA. Data are mean ± SD (n = 3). *p < 0.01. (h) Destabilization of the 3′ UTR lacking the CRE. RAW264.7 cells were co-transfected with pTet-off and either pTRE2-WT or pTRE2-ΔCRE for 18 h, and then treated with 1 μg/ml of doxycycline (Dox) for 2 h. Total RNAs were subjected to qRT-PCR to quantify the 3′ UTR mRNA. Data are mean ± SD (n = 3). *p < 0.01.