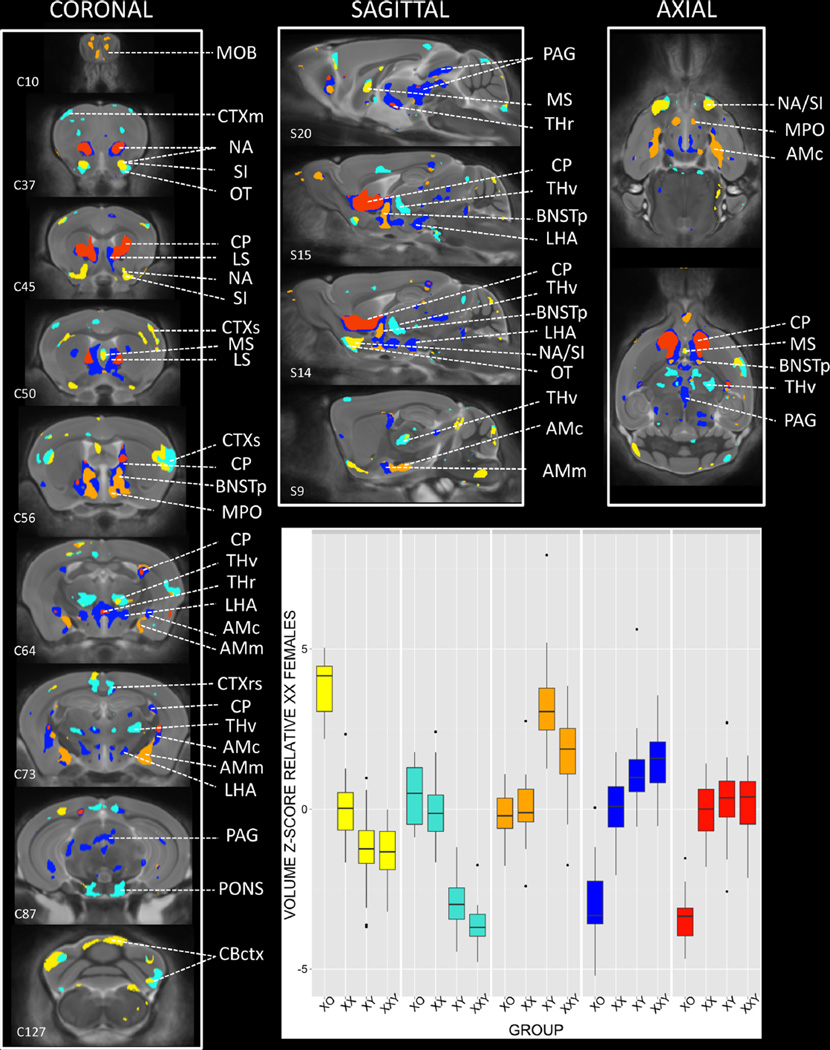
Fig. 3.
Regions of significant anatomical variation across XO, XX, XY, and XXY groups, clustered by observed pattern of anatomical differences between groups. Coronal, sagittal and axial views of the brain show colored regions where an omnibus F test for the effect of group on brain volume survived False Discovery Rate Correction for multiple comparisons with q (expected proportion falsely rejected nulls) set at 0.05. Mean volume for each karyotype group was calculated at every such voxel, and the resulting 200,000 × 4 matrix was subjected to K-means clustering. Visual inspection of the K-means scree plot indicated the optimal number of clusters was 5. Each of these clusters is shown in a different color, with inset box plots displaying the distribution of volume for each cluster across XO, XX, XY, and XXY groups. C# and S# provided for selected slices relate to corresponding Coronal and Sagittal sections within the Allen Institute Reference Mouse Brain Atlas. AMc amygdala, central, AMm amygdala, medial, AON anterior olfactory nucleus, BNSTa anterior division of bed nucleus stria terminalis, BNSTp posterior division of bed nucleus stria terminalis, CP caudate putamen, CBctx cerebellar cortex, CLAUS claustrum, CTXent cerebral cortex: entorhinal, CTXm cerebral cortex: motor, CTXrs cerebral cortex: retrosplenial, CTXss cerebral cortex: somatosensory, HIP hippocampus, LHA lateral hypothalamic area, LS lateral septum, MBret midbrain reticular nucleus, MOB main olfactory bulb, MPO medial preoptic area of hypothalamus, MS medial septal nucleus, NA nucleus accumbens, OT olfactory tubercle, PAG periaqueductal gray, SI substantia innominata, THr Reuniens nucleus of thalamus, THv ventral group nuclei of thalamus