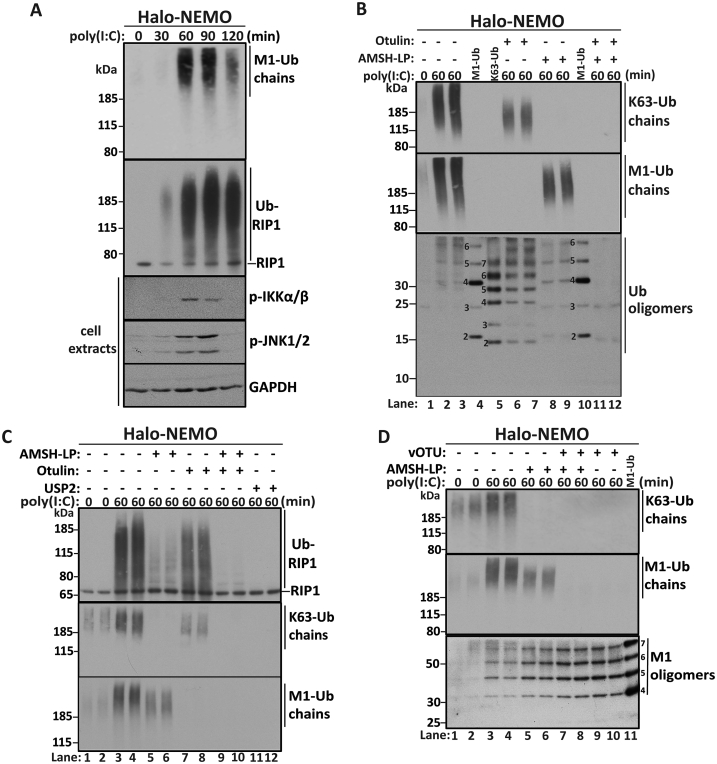
Fig. 4.
Poly(I:C) induces K63/M1-Ub hybrid chain formation in BMDM. (A) BMDM were stimulated with 10 μg/ml poly(I:C) for the times indicated and ubiquitin chains were captured from 2 mg of cell extract protein with Halo-NEMO beads. The captured M1-Ub chains and ubiquitylated RIP1 were identified by immunoblotting. Aliquots of the cell extract (20 μg protein) were subjected to SDS-PAGE and immunoblotting with antibodies that recognize the active phosphorylated forms of IKKα, IKKβ, JNK1 and JNK2, and with GAPDH as a loading control. (B) Similar to (A) except that, after the Halo-NEMO pull-down, samples were treated for 1 h with λ-PPase (100 units) in the absence (−) or presence (+) of Otulin (1.0 μM) and/or AMSH-LP (0.2 μM). The samples were subjected to SDS/PAGE and proteins of >65 kDa immunoblotted for K63-Ub and M1-Ub chains. Proteins <50 kDa were immunoblotted with an antibody that recognizes all forms of ubiquitin to detect the small Ub oligomers released by treatment with DUBs. M1-Ub oligomeric standard marker proteins (12 ng - lanes 4 and 10) and K63-Ub oligomeric standards (5 ng - lane 5) were included to identify the small Ub oligomers formed after treatment with AMSH-LP or Otulin. (C) Similar to B except that, after incubation with Otulin and/or AMSH-LP, the gels were immunoblotted with a RIP1 antibody, as well as with antibodies that recognize M1-Ub and K63-Ub chains. (D) Similar to B, except that the large and small M1-Ub oligomers were detected by immunoblotting before and after treatment with the deubiquitylases AMSH-LP and/or vOTU (0.1 μM).