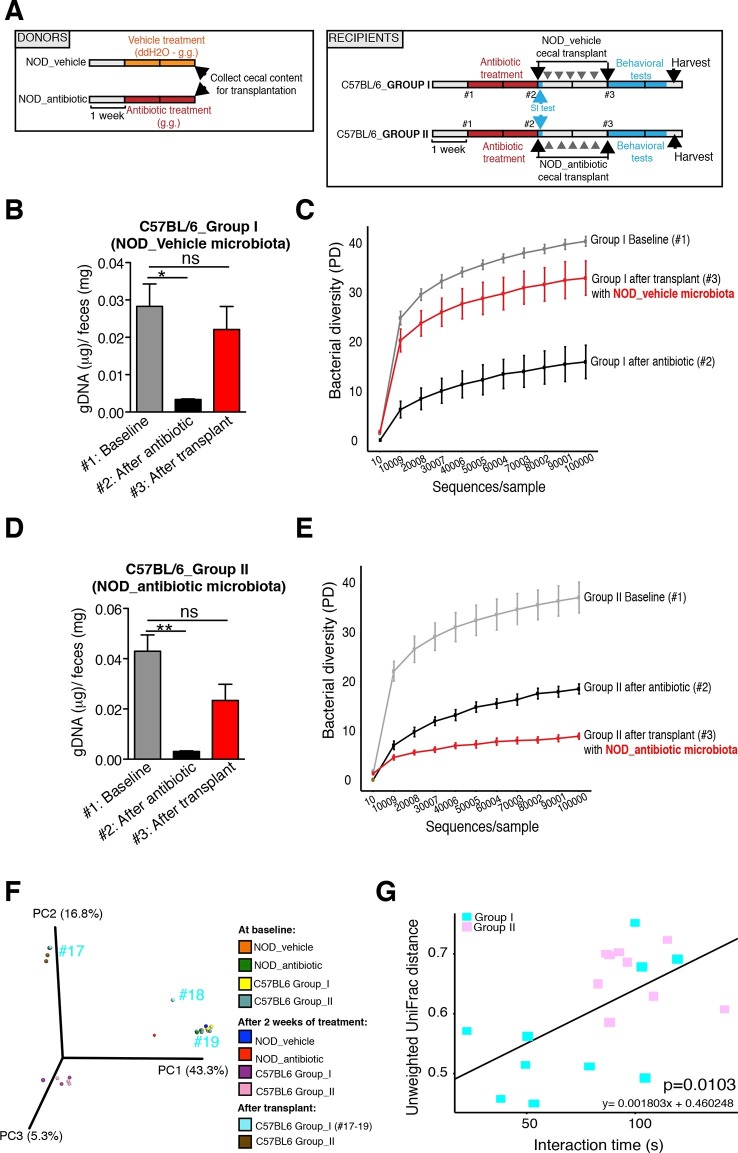
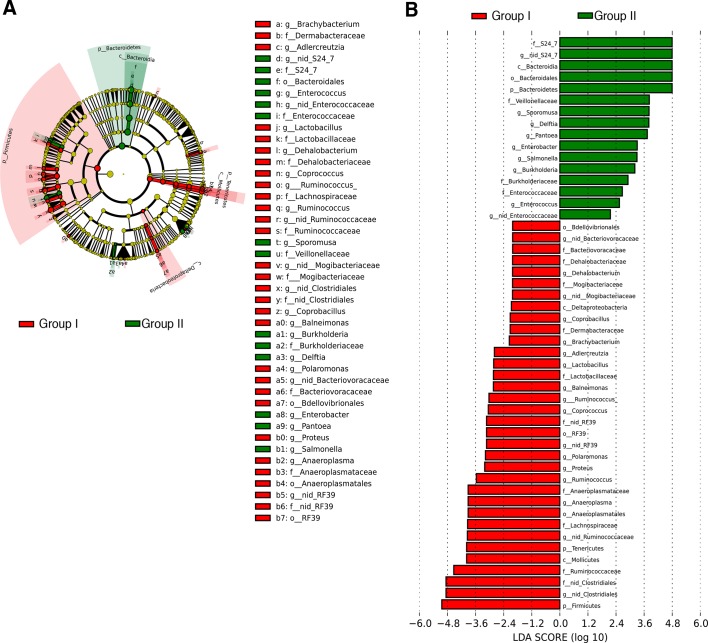
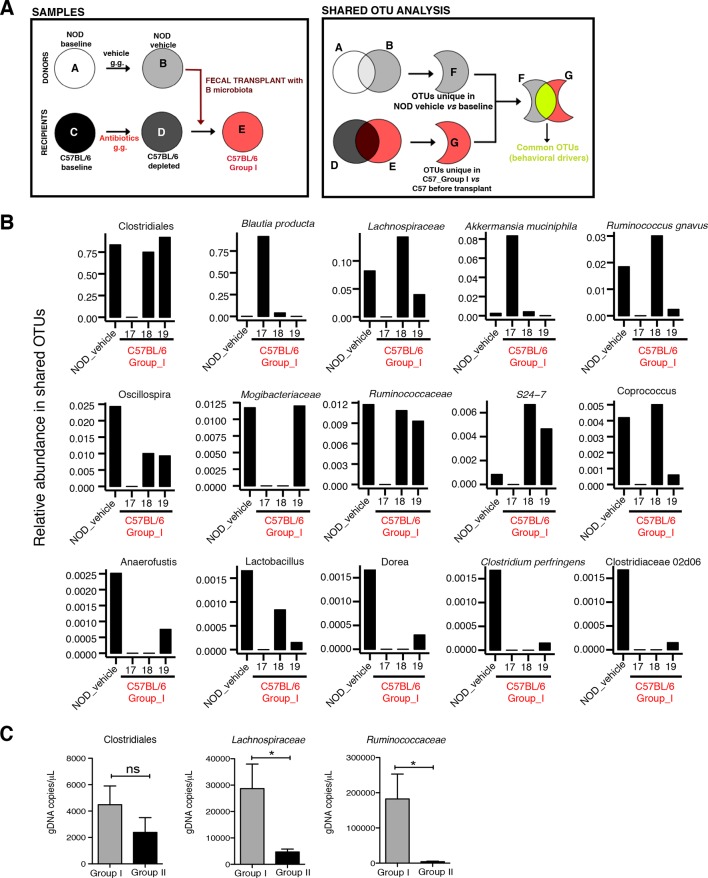
Figure 6. Effect of fecal transplantation on bacterial mass and biodiversity in microbiota depleted C57BL/6 recipients.
(A) Experimental timeline for donors (NOD) and transplanted recipients (C57BL/6). (B,C) Graphs represent fecal biomass (µg of gDNA relative to total fecal weight) of C57BL/6 recipients prior to transplantation (#1 before and #2 after 14 days of antibiotic treatment) and at end point after-transplantation (#3) with donor microbiota (n=3 pooled samples per time-point, each sample represents 1 sample = pooled feces from 3–5 mice. Data are mean ± S.E.M; *p<0.05, **p<0.01 based on one-way ANOVA with Bonferroni’s post hoc test). (C,E) Rarefaction curves comparing alpha diversity of fecal microbiota samples from C57BL/6 recipients at different experimental time-points (#1, #2, and #3). (F) Principal coordinate analysis plot of unweighted UniFrac distances of fecal samples from NOD donors and C57BL/6 mice at different time-points. (#1, #2, and #3). Each dot represents the microbiota of a sample, colored by group, treatment, and time-point (n=3 pooled samples per time-point; each sample corresponds to pooled feces from 3–5 mice). The percentage of variation explained by each principal coordinate (PC) is shown in parentheses. (E) Relationship between social interaction time and unweighted UniFrac distance to NOD donor mice (n=3) for all C57BL/6 recipients (n=10). Each point represents a single C57BL/6 animal, colored by group (light blue: Group_I, transplanted with NOD-vehicle microbiota; pink: Group_II, transplanted with NOD-antibiotic microbiota). Linear regression analysis indicates a significant correlation (p=0.0103) between the variables.