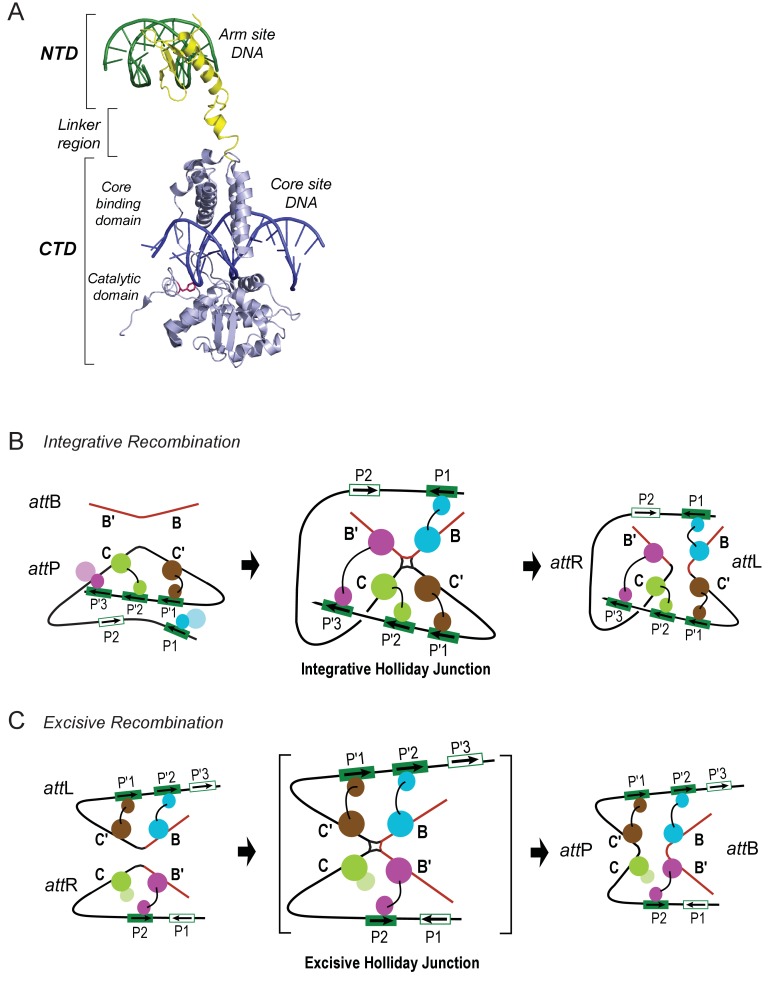
Figure 2. Patterns of specific Int bridges between core- and arm-type DNA binding sites.
(A) The amino-terminal domain (NTD) of Int (yellow) binds to one of the five arm-type DNA sites (green) via a helix-turn-helix recognition motif. It is joined via a flexible 10 residue linker region to the carboxy-terminal domain (CTD) (light blue), which is composed of two smaller domains (the core-binding (CB) and catalytic domains) joined by a 12 residue linker. The CTD binds to core-type DNA sites (blue) via a C-clamp motif; the distal domain of the C-clamp contains the nucleophile Tyr-342 (red) and those residues comprising the catalytic site for DNA cleavage and ligation. The CTD is analogous, and very similar in structure and function, to the well-studied monovalent recombinases, Cre, Flp, and XerC/D. (B) Integrative recombination depends upon four Int bridges, as determined previously (Tong et al., 2014). In the attP and attB substrates (left-most panel) only two of the four bridges are formed prior to synapsis; the accessory DNA bending proteins have been omitted for clarity (see Figure 1C). The coloring scheme for the Ints bound to different arm-type sites is the same as that used in all of the following figures. (C) Excisive recombination involves a different pattern of Int bridges (Tong et al., 2014) and, as shown in Figure 1C, a different ensemble of DNA bending proteins. The HJ intermediate of this reaction (brackets) was purified and used for single particle cryo-electron microscopy, as described in the text.