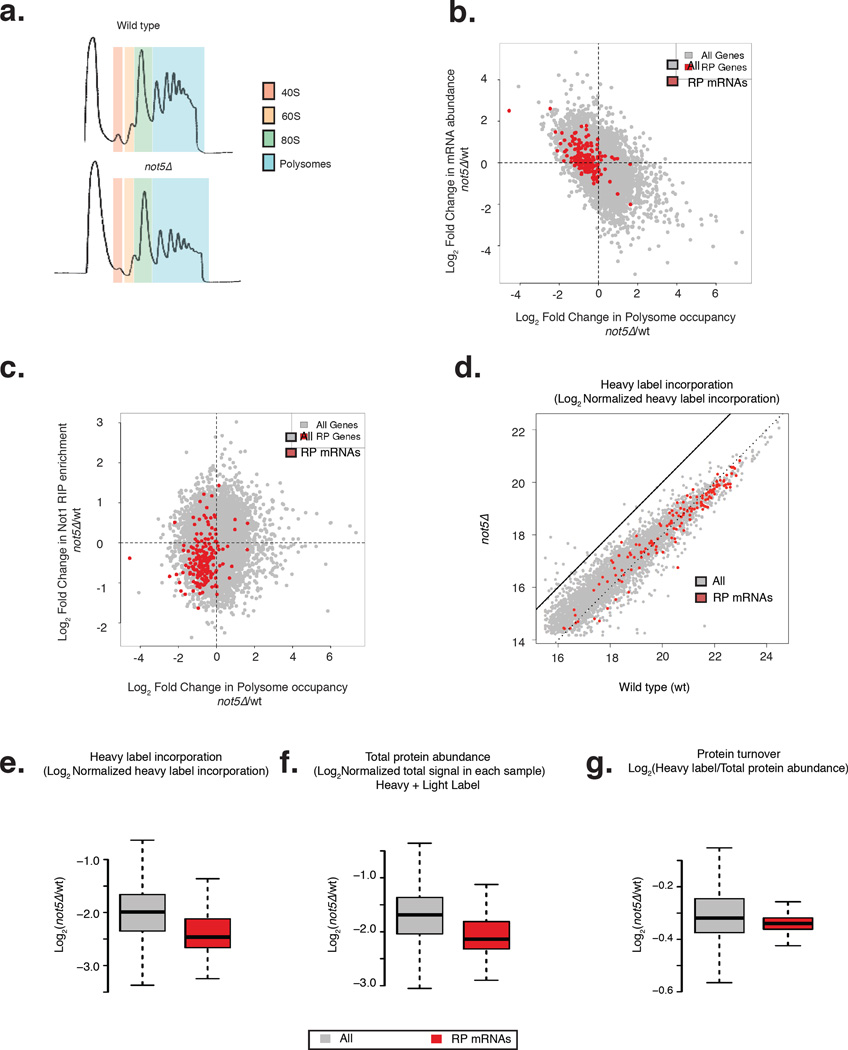
Figure 4. Not5 affects the translation of ribosomal protein genes.
a: Polysome traces in wt cells (top) and not5Δ cells (bottom) with different ribosomal fractions highlighted. b: Scatter plot between Log2 fold change in mRNA abundance from not5Δ to wt against the Log2 fold change in polysomal occupancy from not5Δ to wt. c: Scatter plot between Log2 fold change of Not1 RIP enrichment from not5Δ to wt is plotted against polysomal occupancy change from not5Δ to wt. d: Scatter plot between Log2 transformed heavy label (lysine) incorporation in wt and not5Δ strains from SILAC experiment. e: Box plots for the Log2 fold change in heavy label incorporation from not5Δ to wt. f: Box plots for the Log2 fold change in total protein abundance (sum of both heavy and light lysine) from not5Δ to wild type. g: Box plots for the Log2 fold change in protein turnover (measured by the Log2 ratio of heavy label incorporation over the total protein abundance) from not5Δ to wt. b–g, all mRNAs are in gray and RP mRNAs are in red. See also Tables S2 and S3.