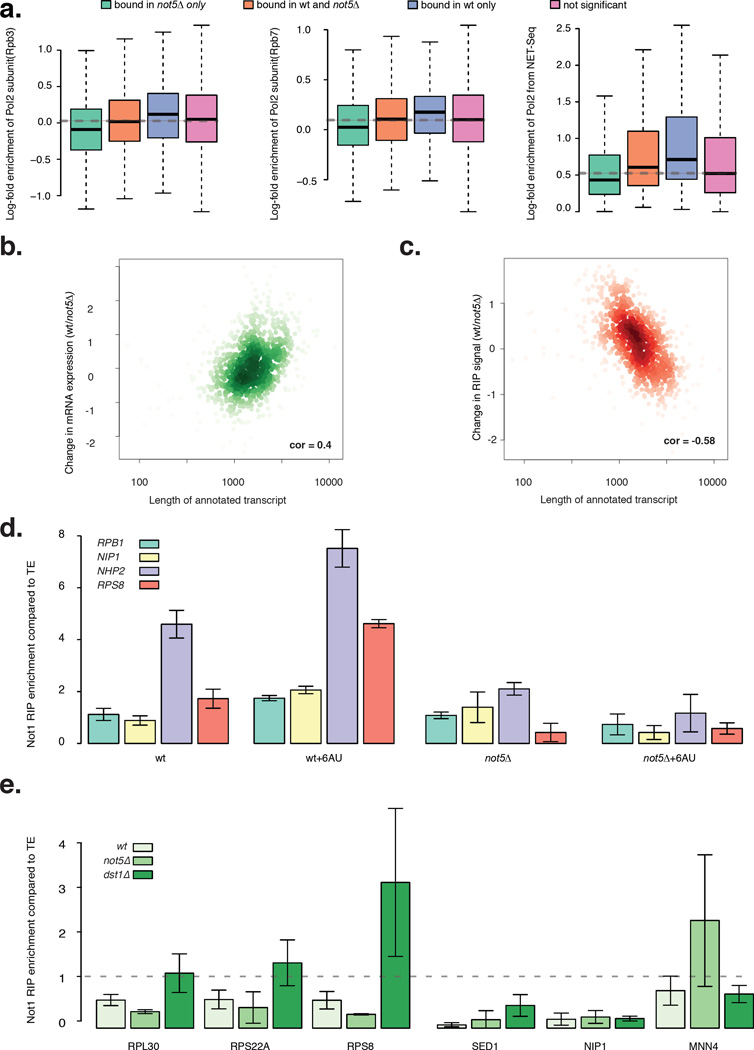
Figure 5. Not5–dependency for Not1 mRNA binding correlates with elongating polymerase.
a: Box-plots show how four different categories of genes categorized by their Not1 binding properties (bound in not5Δ only, bound in wt and not5Δ, bound in wt only and not significantly bound) correlate with polymerase occupancy on genes: Rpb3 (Mayer et al., 2010), Rpb7 (Jasiak et al., 2008) and with Net seq signal identifying nascent mRNAs (Churchman and Weissman, 2011). b: Scatter plot between change in RNA abundance from wt to not5Δ (wt/not5Δ) and transcript length is positively correlated. c: Scatter plot between change in Not1 RIP signal strength (wt/not5Δ) and transcript length is negatively correlated. d: Barplot for Log2 fold change in Not1 enrichment over 4 mRNAs (RPS8, NHP2, RPB1 and NIP1) in wt and not5Δ after transcriptional inhibition upon 6AU treatment. e: Barplot for fold enrichment in Not1 RIP compared to total extract, for several mRNAs in wt, not5Δ and dst1Δ. d–e, error bars representing SD. See also Figures S4, S5 and Table S4.