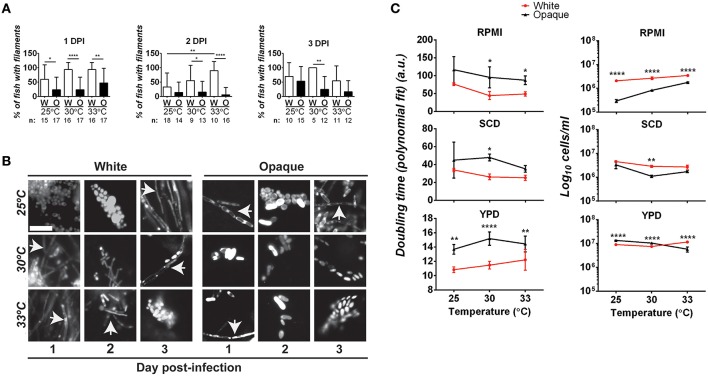
Figure 5.
Comparative analysis of filamentation and growth rates in white and opaque C. albicans cells. (A) Zebrafish larvae were infected with 21–50 AlexaFluor-568 stained C. albicans white (W) or opaque (O) cells expressing dTomato and housed at the indicated temperature after infection. At 1, 2, and 3 days post-infection (DPI) live fish were crushed (see Methods) and assessed for the presence of filaments microscopically. “n” indicates the number of fish examined for each experiment. Shown are the mean percentages of fish containing filamentous C. albicans cells ± SD. *p < 0.05; **p < 0.01; ****p < 0.0001. Data are a compilation of five independent experiments. (B) Representative microscopic images of the fish assessed in (A). Arrows indicate filaments/hyphae. Magnification, 40 ×; scale bar, 20 μm. (C) C. albicans white (CAY4975) and opaque (CAY4986) cells were grown in 96-well plates in YPD, SCD, or RPMI media at 25, 30, or 33°C. Cell growth was analyzed using a Biotek Synergy HT plate reader for 24 h. OD600 was measured at 15 min intervals. Data were analyzed using a previously described Matlab script. Shown are the average Log10 cells/ml at saturation ± SD for white and opaque cells at the designated temperature. Data is representative of 3–4 biological replicates each done in triplicate. Statistically significant differences were determined by two-way ANOVA and Sidak's Multiple Comparisons test. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.