Table 1.
Processing time of Taxonomer compared to rapid classification pipelines SURPI and Kraken. RNA-Seq data generated from three nasopharyngeal specimens with varying taxonomic composition illustrate differences in analysis times between the three tools. (Human: blue; Bacteria: orange; Fungal: green; Virus: red; Other: yellow; Unclassified: gray)
| Sample composition, total reads | Pathogen | Application | Subtraction | Binning | Classification | Protein search | Total time | % Reads classified |
|---|---|---|---|---|---|---|---|---|
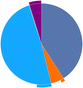
6,599,164 |
HCoV | Taxonomer | - | 5 min | 22 s | 10 s | 5.5 min | 99.9 % |
| Kraken | - | - | 1.5 min | - | 1.5 min | 99.6 % | ||
| SURPI | 3.3 min | - | 74 min | 15 min | 92 min | 99.9 % | ||
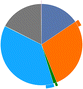
7,542,552 |
Influenza A virus | Taxonomer | - | 8 min | 40 s | 30 s | 9.2 min | 88 % |
| Kraken | - | - | 1.5 min | - | 1.5 min | 66 % | ||
| SURPI | 9.8 min | - | 208 min | 18 min | 236 min | 78 % | ||
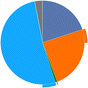
6,252,311 |
HMPV | Taxonomer | - | 5.2 min | 56 s | 10 s | 6.3 min | 98 % |
| Kraken | - | - | 1.3 min | - | 1.3 min | 93 % | ||
| SURPI | 56 min | - | 648 min | 24 min | 728 min | 95 % |
