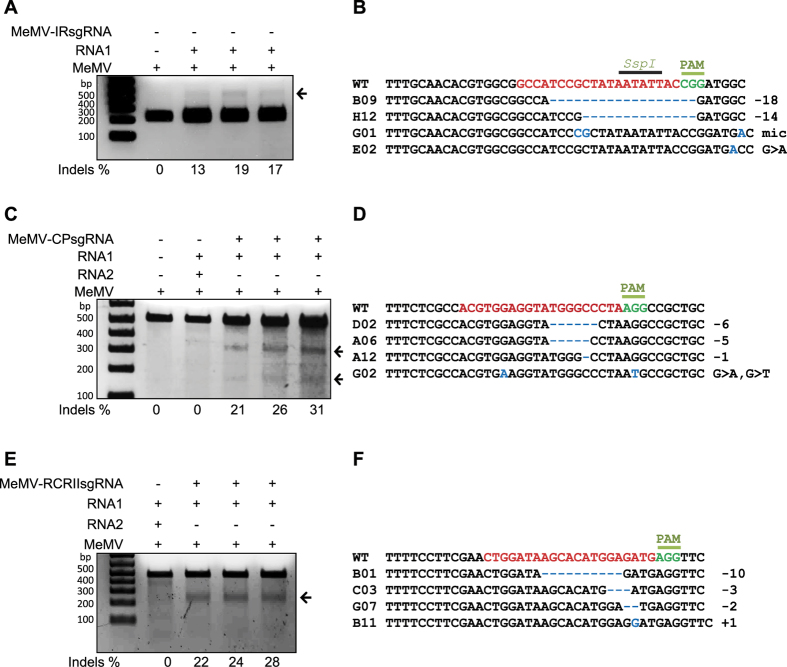
Figure 2. NHEJ repair of coding and non-coding sequences of the MeMV genome.
Non-coding IR, coding CP, and the Rep RCRII domain of MeMV were analyzed for NHEJ repair. (A) NHEJ repair (indel) analysis via an SspI recognition site loss assay. The MeMV IR (453 bp) was analyzed for the loss of the SspI recognition site at the CRISPR/Cas9 target locus. Unlike CLCuKoV, NHEJ-repaired indels are indicated by arrows pointing to the 453-bp SspI-resistant DNA fragments. (B) Alignment of reads of PCR amplicons encompassing the IR of MeMV subjected to Sanger sequencing for NHEJ repair confirmation. (C) T7EI assay to detect indels in the CP of the MeMV genome. The T7EI assay detected indels only in CP PCR amplicons from plants infiltrated with TRV containing the CP-sgRNA, but not in plants infiltrated with TRV empty vector or virus alone. (D) Alignment of reads of the PCR amplicons encompassing the target site and subjected to Sanger sequencing for NHEJ repair confirmation. (E) NHEJ repair analysis at the RCRII motif of MeMV by T7E1 assay. Arrow indicates the expected DNA fragments; TRV empty vector or virus alone did not show similar fragments. All DNA fragments from (A,C and E) were resolved on an 2% agarose gel premixed with ethidium bromide stain. Arrows in (A,C and E) indicate the expected DNA fragments. The indel percentage shown below each gel was calculated based on the Sanger sequence reads. In (B,D and F), the wild-type (WT) sequences are shown at the top (target sequence is shown in red; the protospacer-associated motif [PAM] is indicated in green, followed by the various indels formed, as indicated by numbers to the right of the sequence (−, deletion of x nucleotides; +, insertion of x nucleotides; and >, change of x nucleotides to y nucleotides).