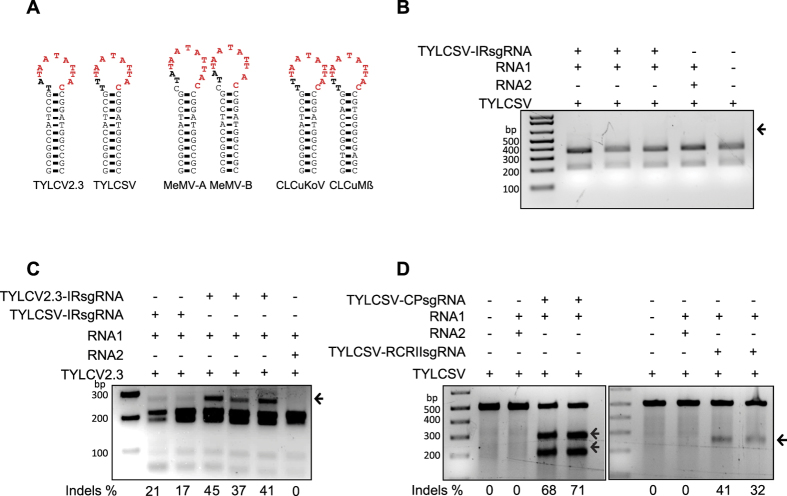
Figure 3. Variable efficiencies of indel formation at the IR sequences of different strains of TYLCV.
(A) Stem-loop structure of the different geminiviruses used in this study. The conserved nonanucleotide motif is flanked on each side by a short stretch of complementary sequences. (B) Restriction site loss assay for detecting NHEJ-based indels at the IR of TYLCSV. The TYLCSV IR (562 bp) was analyzed for the loss of the SspI recognition site at the targeting locus. The arrow indicates the location of the expected 562-bp SspI-resistant DNA fragment in samples with IR-sgRNA, but like TRV empty vector and virus alone, no SspI-resistant fragment was observed. (C) SspI recognition site loss assay for detecting indels at the IR in TYLCV2.3. The variant TYLCSV-IRsgRNA (two lanes after marker) and authentic TYLCV2.3-IRsgRNA (last three lanes) were used to target the IR of TYLCV2.3. Arrows indicate the presence of the expected 269 bp SspI-resistant DNA fragments in samples with TYLCV2.3-IR-sgRNA or TYLCSV-IRsgRNA, but no SspI-resistant fragment was observed in TRV empty vector. (D) T7EI assay for detecting NHEJ-based indels in the CP and RCRII domain of the TYLCSV genome. T7EI assay detected high rates of indel formation both in CP and RCRII PCR amplicons from plants infiltrated with TRV containing CP or RCRII-sgRNA compared with TRV empty vector. DNA fragments of (B,C and D) were resolved on a 2% agarose gel stained with ethidium bromide. Arrows represent the expected DNA fragment in T7EI-digested DNA. The indel percentage shown below each gel was calculated based on the Sanger sequence reads.