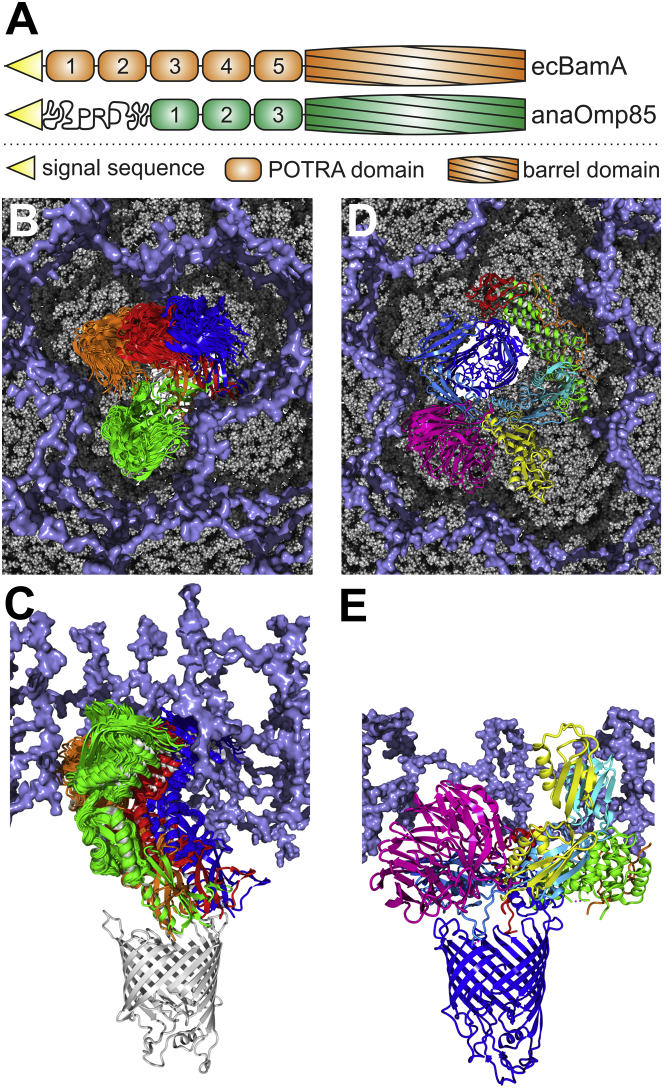
Figure 6.
Model of anaOmp85-POTRA domains embedded in the PGL. (A) The domain compositions of anaOmp85 and ecBamA are shown. PRD denotes the proline-rich domain of anaOmp85. (B and C) An ensemble of MD structures on the outer contour line of the elliptic region in Fig. 5D, representing the most populated orientations of anaP1-anaP2 in MD simulations, and overlapping with the top-scoring Rosetta ensemble, was superimposed onto anaP2 in the crystal structure to indicate the space in which anaP1 rotates relative to anaP2 (Fig. S18). To estimate the rotation of anaP3 relative to the β-barrel, we selected BamA structures that exhibit different orientations of ecP5 relative to the β-barrel (PDB: 4K3B (green), 4K3C (blue), 5AWY (red), and 5EKQ (orange)) and aligned anaP3 onto ecP5. Another structure (PDB: 4C4V) was omitted, because it was a construct including ecP5 and the β-barrel only, which probably resulted in an artificial ecP5 orientation (35). A homology model of the β-barrel of anaOmp85 was built with YASARA based on an alignment with the template structure 4n75 constructed by the HHpred server (80). An ∼10-nm-thick model of the PGL (ice blue) (71) is shown in surface representation. (D and E) The BAM complex (PDB: 5AWY (9)) is shown with its components BamA (domains colored cyan to blue from N- to C-terminus), BamB (magenta), BamC (orange), BamD (green), and BamE (red). For ecP1 and ecP2, an additional conformation (PDB: 3EFC) is shown, which exhibits a different angle at the switch region between ecP2 and ecP3 (yellow). The PGL was cut to match the reported thickness of 5–6 nm in E. coli. The models in (B) and (D) are not shown on the same scale.