Figure 1.
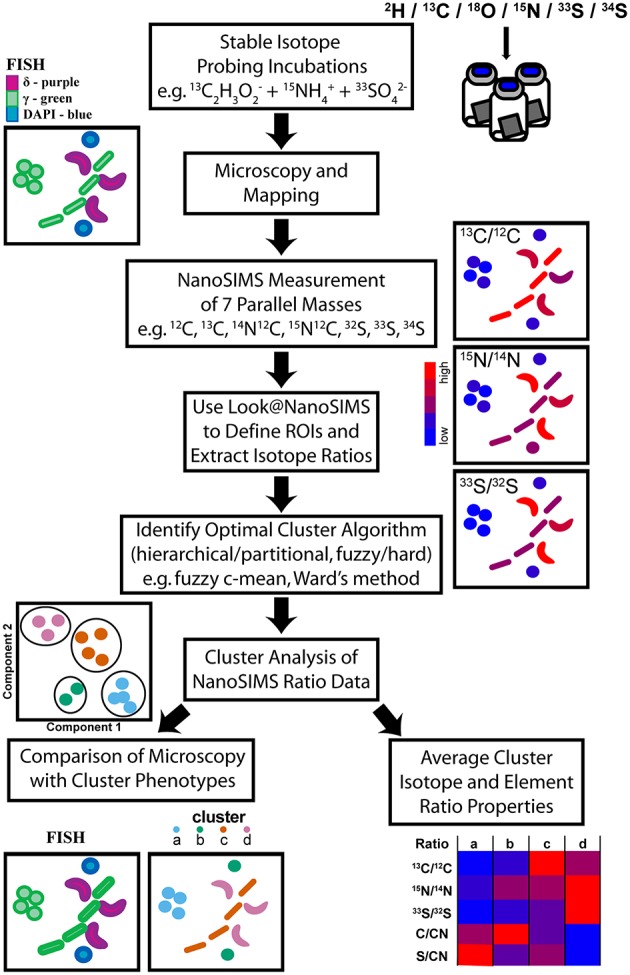
Illustration of the workflow from multiple-isotope SIP incubations through to the identification of stable isotope phenotypes and their properties via cluster analysis of NanoSIMS data. After colonization with microbial biomass, NanoSIMS compatible Si-wafers were incubated with multiple stable isotope labeled substrates. Microscopy, using FISH probes, identified regions that were mapped for subsequent NanoSIMS analysis of up to seven parallel masses. NanoSIMS data was processed using Look@NanoSIMS to define single cell regions of interest (ROIs) and extract associated isotope and elemental composition ratios. Several cluster analysis algorithms were evaluated to determine the best method, which was then applied to partition the NanoSIMS ratio data. The properties of the stable isotope phenotype clusters were examined and compared to independent FISH images to investigate label uptake in a multispecies metabolic network.
