Figure 1. Application of the CD Method to Antidromically Driven In Vitro Spinal Cord Data.
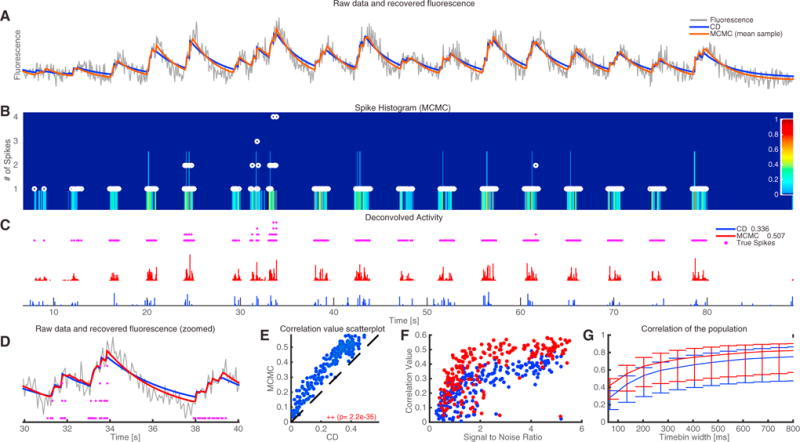
(A) Raw fluorescence data from an example neuron (gray) and reconstructed fluorescence trace with the proposed CD method (blue) and the mean sample obtained by the fully Bayesian MCMC method of Pnevmatikakis et al. (2013), with time constant updating (red). The CD method effectively denoises the observed fluorescence trace but overestimates the time constants slightly, while the more expensive MCMC method fine-tunes the time constants to match better the observed data.
(B) Color-coded depiction of the empirical posterior marginal histogram obtained with the MCMC method and true number of antidromic spikes during each timebin (white dots). The colormap displays the probability of a certain number of spikes within a given timebin. The MCMC method can quantify uncertainty and identify multiple spikes within a single timebin.
(C) Estimated neural activity (normalized) from the CD method (blue) and mean of the posterior marginal per timebin with the MCMC method (red). The legend also shows the spike correlation for each method at the imaged resolution. All methods detect accurately the bursting intervals of the neurons. The more expensive MCMC method gives a significant improvement in the spike deconvolution according to the spike correlation metric.
(D) Zoomed-in version of (A).
(E) MCMC outperforms CD for this dataset (Wilcoxon signed ranked test). Each circle corresponds to a single cell.
(F) Correlation values at the imaged resolution for all n = 207 cells as a function of the signal-to-noise ratio for the two methods. Performance increases with the SNR for all methods. Again, each circle corresponds to a single cell.
(G) Median correlation values for all n = 207 cells at various timebin widths. Error bars indicate the 0.25 and 0.75 quantiles, respectively.
