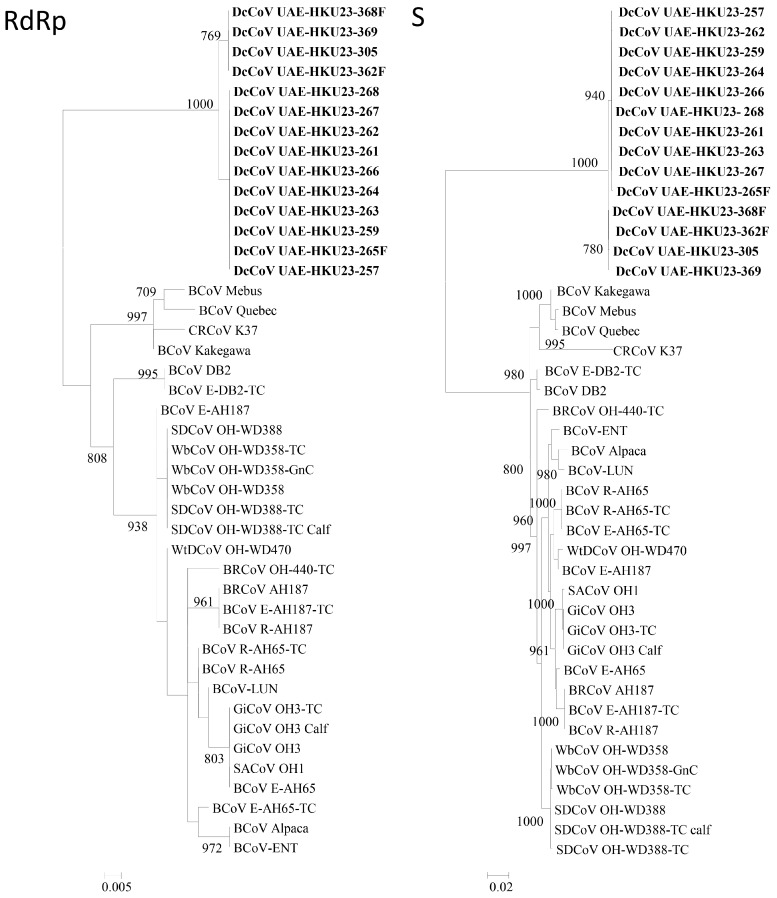
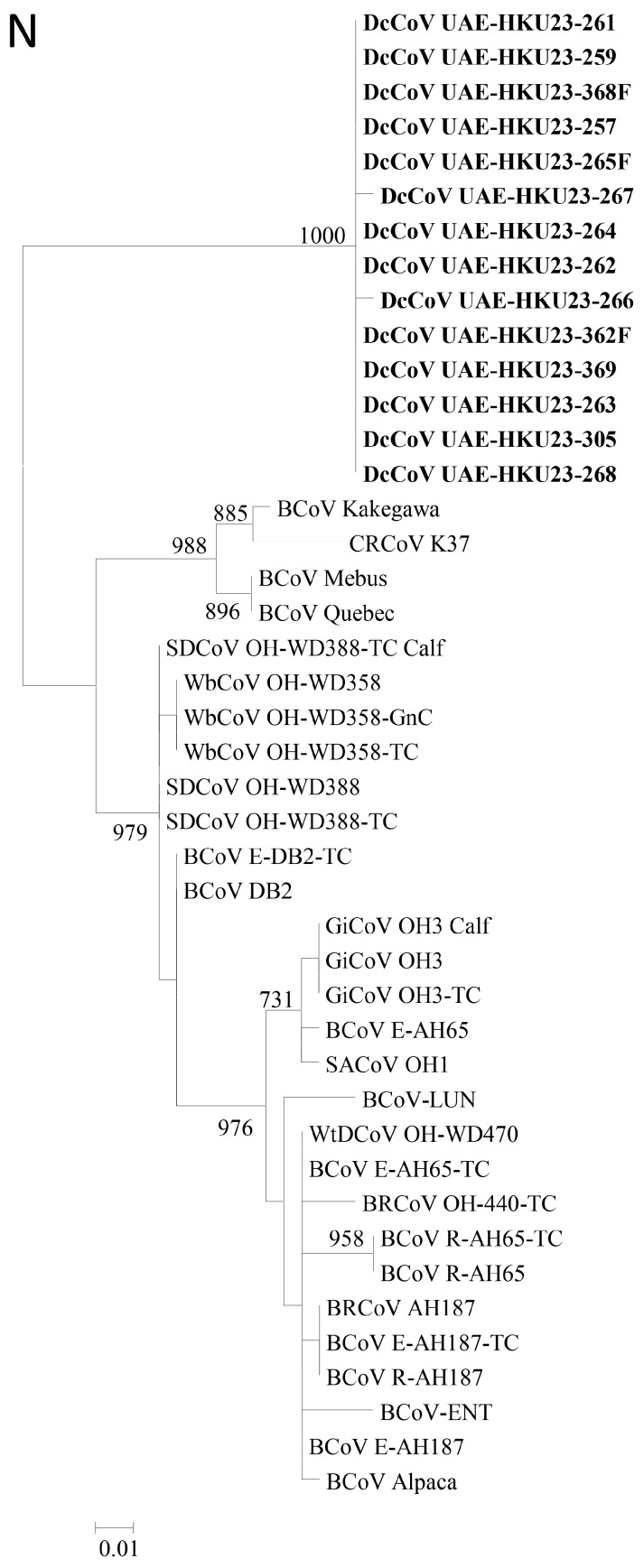
Figure 5.
Phylogenetic analyses of RNA-dependent RNA polymerase (RdRp), S, and N genes of dromedary camel coronavirus (DcCoV) UAE-HKU23. Included in the analysis were 2784, 4101, and 1347 nucleotide positions in RdRp, S and N, respectively. For RdRp, the scale bar indicates the estimated number of substitutions per 200 nucleotides. For S, the scale bars indicate the estimated number of substitutions per 50 nucleotides. For N, the scale bars indicate the estimated number of substitutions per 100 nucleotides. Bootstrap values were calculated from 1000 trees and those below 70% are not shown. The 14 strains of DcCoV UAE-HKU23 characterized in this and our previous studies are shown in bold. BCoV, bovine coronavirus; CRCoV, canine respiratory coronavirus; SDCoV, sambar deer coronavirus; WbCoV, waterbuck coronavirus; WtDCoV, white-tailed deer coronavirus; BRCoV, bovine respiratory coronavirus; GiCoV, giraffe coronavirus; SACoV, sable antelope coronavirus.