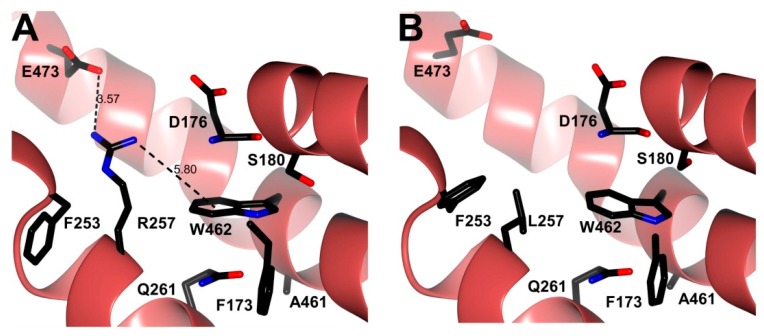
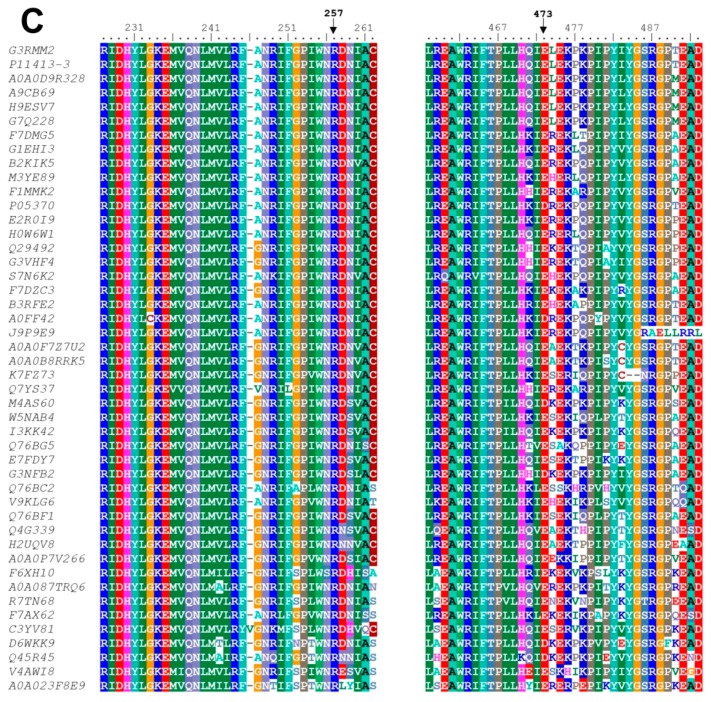
Figure 9.
Structural comparison between the human WT G6PD enzyme and the Class I G6PD Zacatecas mutant. (A) Crystallographic structure of human WT-G6PD enzyme (pale crimson) (PDB entry 2BH9); (B) Minimized model of the Class I G6PD Zacatecas variant (pale crimson) with the in silico R257L mutation. Note that the R257 (black cylinders) residue forms a weak cation–π interaction with W462 and a salt bridge with E473 in (A), and their absence in the in silico R257L mutant in (B). Distances are in Å; (C) Alignment of amino acid sequence of G6PD with homologs from different species from Gorilla gorilla gorilla (G3RMM2), Homo sapiens (P11413-3), Chlorocebus sabaeus (A0A0D9R328), Papio anubis (A9CB69), Macaca mulatta (H9ESV7), Macaca fascicularis (G7Q228), Equus caballus (F7DMG5), Camelus dromedarius (G1EHI3), Rhinolophus ferrumequinum (B2KIK5), Mustela putorius furo (M3YE89), Bos taurus (F1MMK2), Rattus norvegicus (P05370), Canis lupus familiaris (E2R0I9), Cavia porcellus (H0W6W1), Macropus robustus (Q29492), Sarcophilus harrisii (G3VHF4), Myotis brandtii (S7N6K2), Ornithorhynchus anatinus (F7DZC3), Sorex araneus (B3RFE2), Mus caroli (A0FF42), Canis lupus familiaris (J9P9E9), Crotalus adamanteus (A0A0F7Z7U2), Pelodiscus sinensis (K7FZ73), Bos indicus (Q7YS37), Xiphophorus maculatus (M4AS60), Lepisosteus oculatus (W5NAB4), Oreochromis niloticus (I3KK42), Ambystoma mexicanum (Q76BG5), Danio rerio (E7FDY7), Gasterosteus aculeatus (G3NFB2), Cephaloscyllium umbratile (Q76BC2), Callorhinchus mili (V9KLG6), Lepisosteus osseus (Q76BF1), Rhabdosargus sarba (Q4G339), Takifugu rubripes (H2UQV8), Scleropages formosus (A0A0P7V266), Xenopus tropicalis (F6XH10), Stegodyphus mimosarum (A0A087TRQ6), Capitella teleta (R7TN68), Ciona intestinalis (F7AX62), Branchiostoma floridae (C3YV81), Tribolium castaneum (D6WKK9), Rhipicephalus microplus (Q45R45), Lottia gigantea (V4AWI8), Triatoma infestans (A0A023F8E9), Anopheles gambiae (H2KMF3), Apis mellifera (A0A023FG14), Musca domestica (T1PD22), Aedes aegypti (Q0IEL8) performed with BioEdit V.7.2.5. The uniform colors indicate conserved amino acid in the sequences reported. Colorless represent non-conserved amino acid sequences. The arrows indicate the position R257 and E473 residues are highly conserved in different organisms.