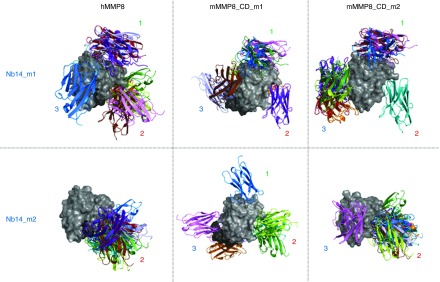
Figure 4.
In silico docking models of the interaction between mMMP8_CD and hMMP8_CD with Nb14. Docking models were obtained for the best Nb14 models (Nb14_m1 and Nb14_m2, in color) in combination with hMMP8_CD (gray) on one hand and the two best models for mMMP8_CD (mMMP8_CD_m1 and mMMP8_CD_m2, gray) on the other hand. These simulations show three possible binding sites (1–3). Two (1 and 3) of which are present outside the active site and one (2) at the active site. Modeling of hMMP8_CD was based on the experimental structure, while homology models (mMMP8m1 and Swissmodel mMMP8) were built for mMMP8_CD. Note the extra linker for mMMP8m1 (dark gray). Multiple templates (PDB: 4LAJ, 3EZJ, 3TPK and 4M3J) were used to construct Nb14, depicted as MMP8_Nbm1 and MMP8_Nbm9. All models were validated by RAMPAGE46 and the best models were used for docking by ClusPro47 to predict binding of the MMP8_Nb to MMP8. Homology models and docking results were analyzed and figures rendered using PyMOL. CD, catalytic domain; h, human; m, mouse; MMP, matrix metalloproteinase; Nb, nanobody.