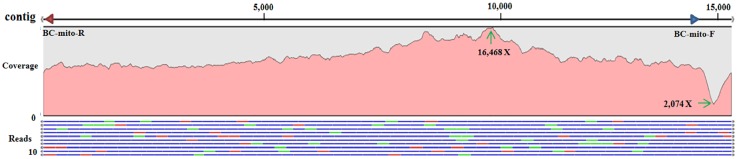
Fig 1. A schematic representation of de novo assembling and coverage estimate of the 15,263 bp mitogenome of Bactericera cockerelli using MiSeq data.
In the top Contig line, the blue and red arrows represent the forward and reversed primers (BC-mito-F/BC-mito-R) used to verify the circularity of B. cockerelli mitogenome by PCR. Numbers are nucleotides in bp. In the Coverage section, the pink area represents nucleotide coverage with the highest of 16,468 X in nad6 and the lowest of 2,074 X in CR. In the Reads section, ten top read assemblings from MiSeq data were representatively shown with blue as pair reads, red as forward reads only and green as reversed reads only.