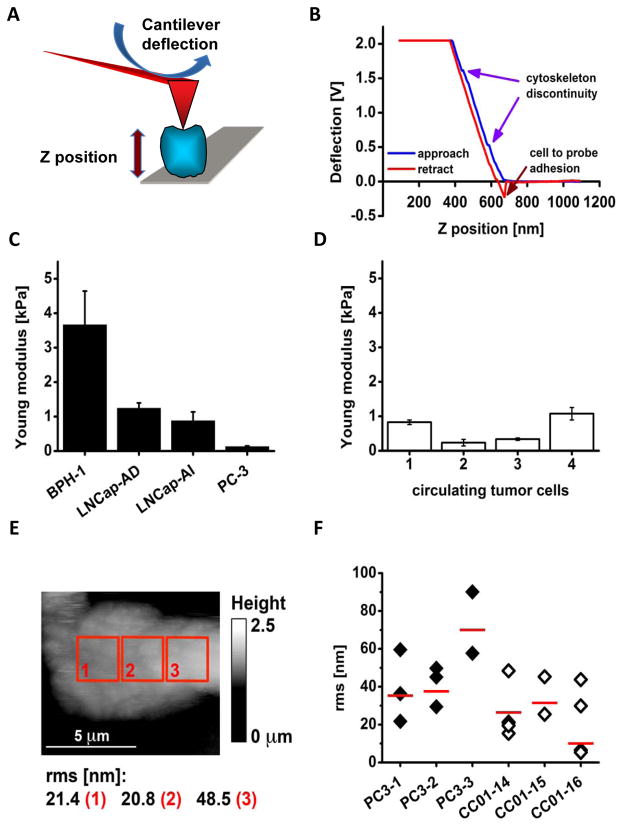
Fig. 2.
AFM probing of cell surface indicates that CTCs exhibit mechanical phenotype resembling highly metastatic cultured prostate cancer cells. A: A scheme illustrating the principle of measuring cell elasticity. A cell (blue) bound to a mica surface (grey) is indented by a tip (red triangle) mounted on a flexible cantilever (red board) proportionally to the cell elasticity. Deflection of the cantilever (blue arrow) changes a position of a laser beam reflection that measures force needed to indent the cell. The distance between a tip end and the cell is represented by the Z position (thick vertical arrow) directly measured by a piezoelectric element of the microscope. B: An example of a force plot of individual CTC (cell #4). Blue arrows point at positions of little humps at which the tip likely sensed a cytoskeleton discontinuity. Adhesion forces between the tip and the cell bent the cantilever in the opposite direction as indicated by the red arrow. C: Histogram comparing elasticity of four prostate cell lines. The elasticity is presented as the Young modulus. The benign BPH-1 cells are the stiffest (showed the largest Young modulus), whereas androgen dependent LNCap are more elastic followed by the LNCap androgen independent, and by PC-3 cells that are highly metastatic, and also the softest. Histograms represent mean values with corresponding SD. D: Histogram comparing elasticity of four individual CTCs. These cells were as soft as the cancerous cell lines presented in the panel C. Histograms represent mean values with corresponding SD. E: Height topography image of a single CTC (cell #4) recorded with a spherical tip in contact mode. Roughness (rms in nm) of the cell membrane calculated for three 2.5 by 2.5 μm. The cell is flat since it is tightly bound to a glass plate with poly-L-Lys and also may represent a strongly metastatic phenotype.