Extended Figure 4.
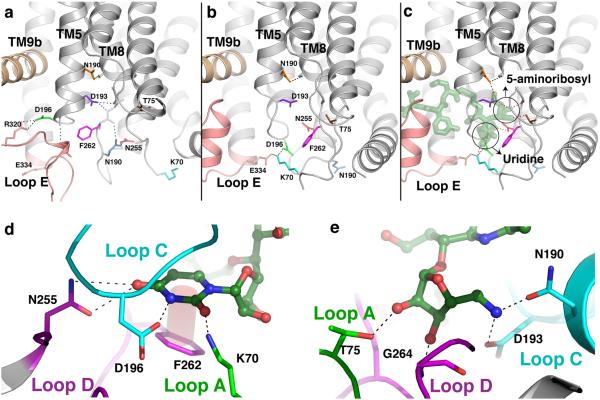
Conformational changes in MraYAA that create binding pockets for the uridine and 5-amino ribosyl groups of MD2. a, A close-up view of apoMraYAA with key residues that participate in conformational changes upon MD2 binding shown as sticks in various colors. b, A close-up view of the nucleoside-binding pocket in the MraYAA-MD2 complex with MD2 omitted. Key residues are colored as in (a). c, A close-up view of the interactions MD2 (green) makes with the nucleoside-binding pocket of MraYAA. Interactions between MraYAA and MD2 are shown as dotted lines. It is noteworthy that residues interacting with the uridine moiety of MD2 move large distances (5-17 Å for residues K70, D196, N255, and F262), while the residues binding the 5-aminoribosyl group of MD2 (T75, N190, and D193) do not make large sidechain movements upon MD2 binding. The uridine and 5-amino ribosyl groups of MD2 are circled. d, Interactions between the uracil base of MD2 (green) and the nucleoside-binding pocket of MraYAA. The uracil base forms H-bonds with side chains of N255, D196 and K70 and forms a π - π interaction with F262. d, The 5-amino ribosyl group of MD2 forms H-bond interactions with side chains of T75, N190, and D193, and the backbone amide of G264.