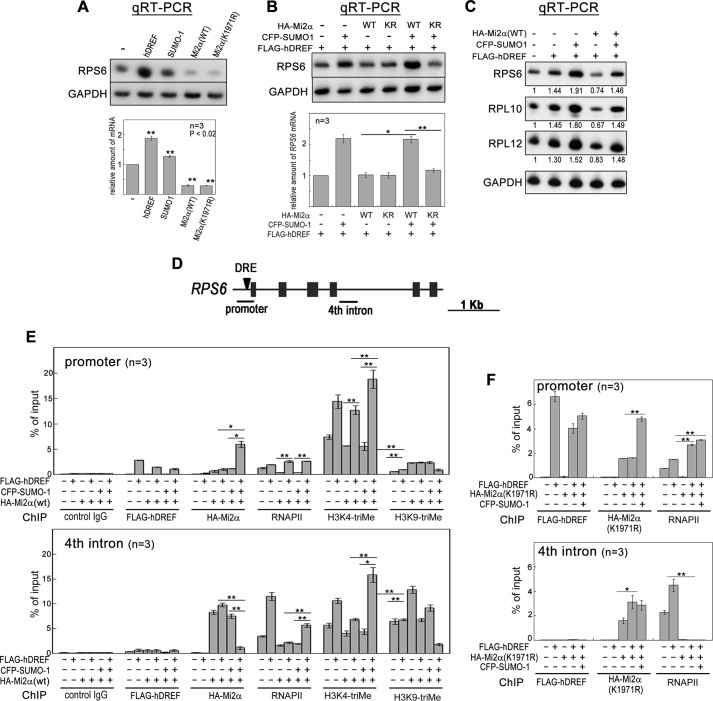
FIGURE 7.
hDREF and SUMO-1 coexpression overcomes transcriptional repression by Mi2α. A, HeLa cells were transfected with plasmid expressing FLAG-hDREF, HA-Mi2α (WT or K1971R), or CFP-SUMO-1 as indicated. At 48 h after transfection, total RNA was extracted, and qRT-PCR was performed for RPS6 and GAPDH. The intensity of individual bands was quantified and expressed relative to the control (no transfection). Amounts of RPS6 mRNA were normalized to those of GAPDH as an internal control. Data are the mean of three independent experiments. Error bars, S.E. **, p < 0.02; n = 3. B, HeLa cells were transfected with FLAG-hDREF, HA-Mi2α (WT or K1971R), or CFP-SUMO-1 in the indicated combinations. At 48 h after transfection, total RNA was extracted, and qRT-PCR was performed for RPS6 and GAPDH. Amounts of RPS6 mRNA were normalized by those of GAPDH as an internal control. Data are the average of three independent experiments. Error bars, S.E. *, p < 0.05; **, p < 0.02; n = 3. C, HeLa cells were transfected with FLAG-hDREF, HA-Mi2α (WT or K1971R), and CFP-SUMO-1 as indicated. At 48 h after transfection, total RNA was extracted, and qRT-PCR was performed for RPS6, RPL10, RPL12, and GAPDH. Amounts of mRNA of RPS6, RPL10, and RPL12 were normalized by those of GAPDH as an internal control. Data are expressed as the average value relative to a no transfection control. n = 3. D, schematic illustration of the RPS6 gene. Arrowhead, a hDREF binding site. Solid bars, the region amplified by PCR in ChIP analysis. E, 293FT cells were transfected with FLAG-hDREF, HA-Mi2α, and CFP-SUMO-1 as indicated. At 36 h after transfection, cells were cross-linked with 1.1% formaldehyde for 5 min, lysed, and sonicated. After centrifugation, the cleared supernatant containing chromatin was subjected to immunoprecipitation using antibodies (control IgG, anti-FLAG, anti-HA, anti-RNA polymerase II (phospho-CTD), anti-histone H3 Lys-4-trimethyl, and anti-histone H3 Lys-9-trimethyl). PCR was quantitatively performed in the reaction mixture containing 32P-dCTP using genomic DNA from the input extract and DNA recovered from the immunoprecipitates as indicated. PCR products were separated by electrophoresis on 6% acrylamide gels and detected by autoradiography. The intensity of individual bands was quantified and expressed as a percentage of input. Data are the average of three independent experiments. Error bars, S.E. (*, p < 0.05; **, p < 0.02). F, ChIP assays were performed using antibodies as described. Instead of WT Mi2α, SUMOylation-defective K1971R mutant was expressed in 293FT cells. Data are the average of three independent experiments. Error bars, S.E. (*, p < 0.05; **, p < 0.02).