Figure 2.
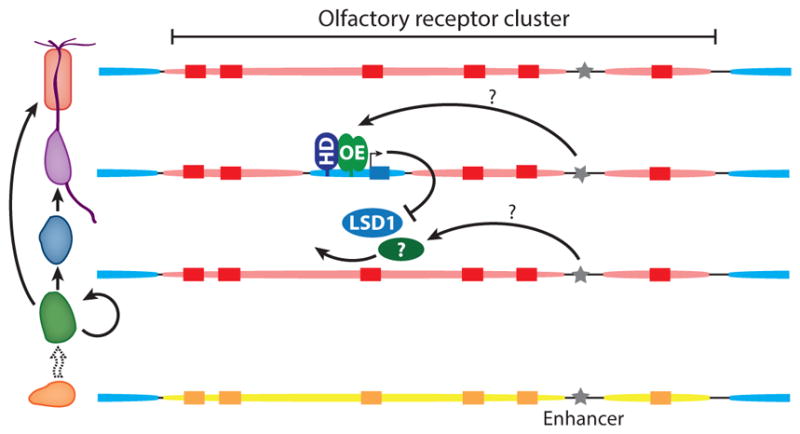
Epigenetic regulation of olfactory receptor (OR) genes. OR genes (rectangles) are embedded within cluster-wide chromatin domains. In horizontal basal cells, OR clusters are decorated with histone modifications ( yellow) characteristic of facultative heterochromatin (H3K9me2 and H4K20me2); flanking euchromatin is blue. In sustentacular cells and immature olfactory sensory neurons, OR clusters bear marks of constitutive heterochromatin (H3K9me3 and H4K20me3) (red ovals). OR activation involves the derepression of a single OR by the histone demethylase LSD1 and additional, unidentified chromatin-modifying enzymes. Following derepression, OR transcription is induced by the binding of O/E family (OE) and homeodomain (HD) transcription factors to OR promoters. OR transcription triggers a negative feedback mechanism that prevents derepression of additional ORs by downregulating LSD1. OR enhancer elements ( gray stars) are euchromatic in main olfactory epithelium tissue as a whole, but their chromatin state has not been studied in individual cell types. OR enhancers could contribute to OR activation by targeting OR derepression, by activating OR transcription following derepression, or both.
