Fig. 1.
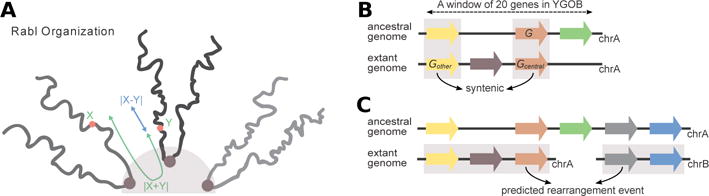
Computational procedures applied to demonstrate the impact of spatial alignment of chromosomal arms on rearrangement frequency. (A) |X − Y| as a measure of spatial alignment between genes located on different chromosomes in the Rabl-like configuration, where X and Y are the genomic distances between these genes and centromeres. (B) A scheme illustrating the inference of synteny. A table of homologous genes centered on the ancestral gene G is downloaded from YGOB. Homology pillars are highlighted in gray. Genes Gcentral and Gother are assumed to be syntenic if they are located within 20 genes from each other in both the extant and ancestral genomes. (C) Prediction of rearrangements as events between two synteny blocks (highlighted in gray) located at different chromosomes in the extant yeast genome, and at the same chromosome within 20 genes from each other in the ancestral genome.
