Figure 7. Potential Allosteric Residues Add a Layer of Annotation to Structures in the Context of Disease-Associated SNVs.
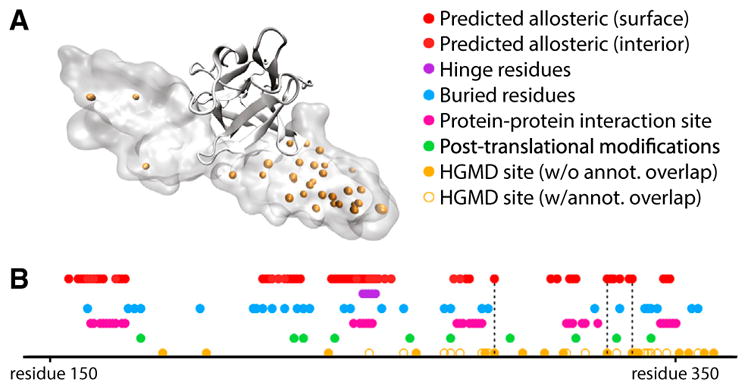
(A) Structure of the fibroblast growth factor receptor (FGFR) in VMD Surf rendering, with HGMD SNVs shown in orange, bound to FGF2, in ribbon rendering (PDB: 1IIL).
(B) Linear representation of structural annotation for FGFR. Dotted lines highlight loci which correspond to HGMD sites that coincide with critical residues, but for which other annotations fail to coincide. Deeply buried residues are defined to be those that exhibit a relative solvent-exposed surface area of 5% or less, and binding-site residues are defined as those for which at least one heavy atom falls within 4.5 Å of any heavy atom in the binding partner (heparin-binding growth factor 2). The loci of post-translational modification sites were taken from UniProt (UniProt: P21802). See also Figures S5 and S6.
