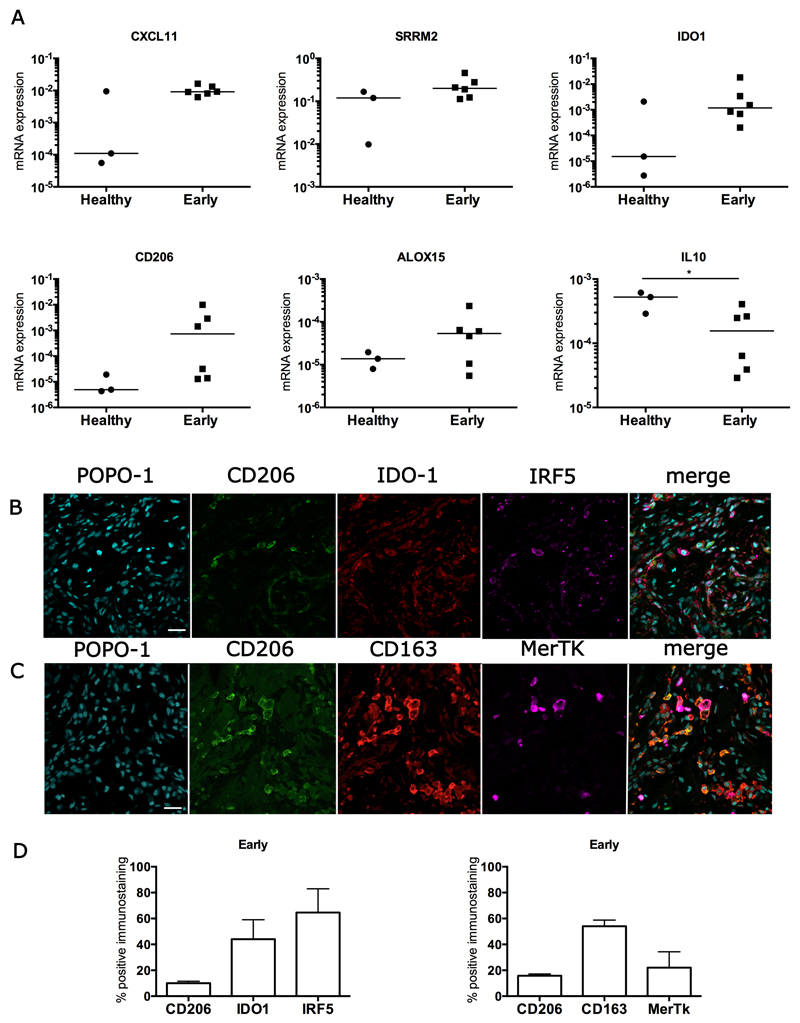
Figure 2.
Expression of inflammation activation pathway genes and proteins in early stage diseased supraspinatus tendons compared to healthy subscapularis tendons. (A) A mixed gene expression signature was present in pre-treatment tendon samples with early disease (n=6) compared to healthy subscapularis control tendons (n=3). Shown is expression of genes induced by IFN including CXCL11, and Serine/arginine repetitive matrix 2 (SRRM2); induced by NF-κB including Indoleamine 2,3-dioxygenase 1 (IDO1) and induced by STAT-6 ALOX15 and CD206. Fold changes in gene expression are shown in Table S2. There was greater expression of IFN-induced genes including CXCL11 and SRRM2, and the NF-κB-induced gene IDO1 in early stage disease supraspinatus tendons compared to healthy subscapularis control tendons. Gene expression is normalized to β-actin; bar shows median value. (B, C) Representative immunofluorescence images of sections of pre-treatment early stage disease tendons stained for inflammation activation markers including those in the STAT-6 pathway (CD206, green), the glucocorticoid receptor pathway (CD163 red), the IFN pathway (IRF5, purple) and the NF-κB pathway (IDO-1, red). MerTK (purple) represents Mer tyrosine kinase, a tissue resident macrophage marker. Cyan represents POPO-1 nuclear counterstain. Scale bar = 20µm. (D) Quantitative analysis showing percentage of immunopositive staining for inflammation activation markers. Data are shown as mean and SEM.