Figure 3.
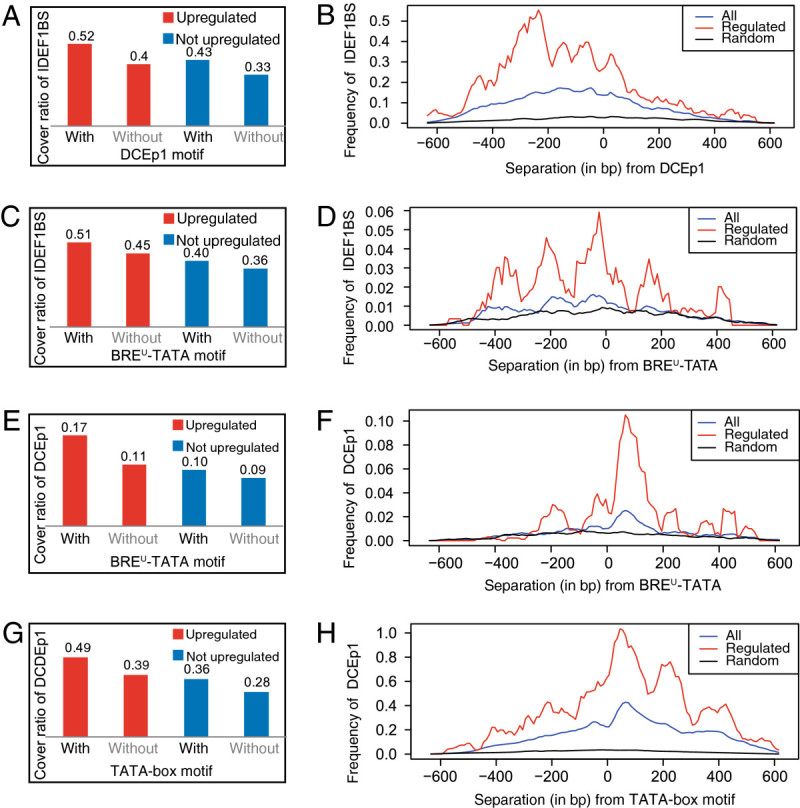
Separations (in bp) between motifs extracted by MAMA. A, Cover ratio of the IDEF1BS motif with (CR(IDEF1BS|DCEp1|UP), CR(IDEF1BS|DCEp1|!UP)) and without the DCEp1 motif (CR(IDEF1BS|!DCEp1|UP), CR(IDEF1BS|!DCEp1|!UP)). B, Separations between the IDEF1BS and DCEp1 motifs. Separations between the IDEF1BS and DCEp1 motifs were calculated, and the number of IDEF1BS motifs co-localizing with DCEp1 motifs in a region 500 bp upstream and 150 bp downstream of the TSSs was counted. Black lines, frequency of motifs in random and 650-bp sequences; blue lines, number of motifs in all genes; red lines, number of motifs in genes upregulated over twofold in response to iron deficiency. Frequency represents the number of IDEF1BS motifs in each ±25-bp window in random sequences, upregulated genes, and all genes divided by the number of random sequences (31,348), upregulated genes (895), and all genes (31,348). C, Cover ratio of the IDEF1BS motif with the BREU-TATA motif 1. D, Separations between the IDEF1BS motif and BREU-TATA motif 1. E, Cover ratio of the DCEp1 motif with the BREU-TATA motif 1. F, Separations between the DCEp1 motif and BREU-TATA motif 1. G, Cover ratio of the DCEp1 motif with the TATA-box motif. H, Separations between the DCEp1 motif and TATA-box motif.
