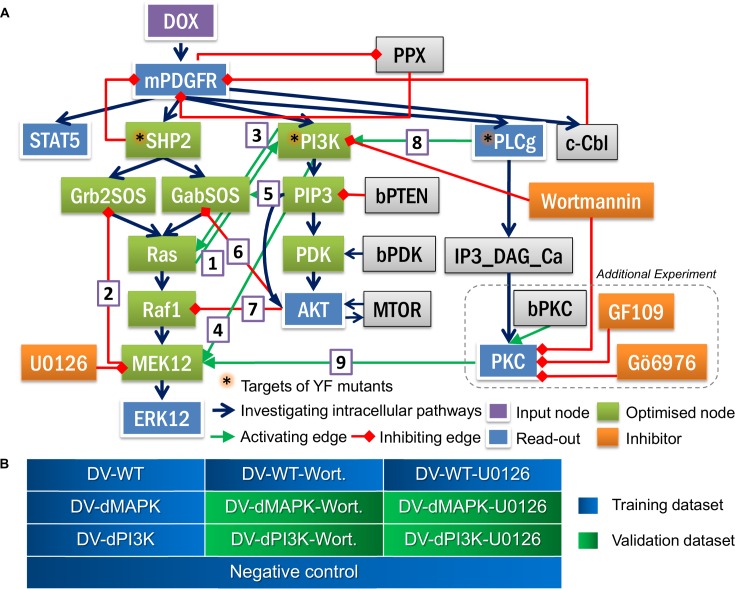
Fig 3. Model structure and datasets for modelling.
[A] The model structure of the literature-derived PDGF signalling is shown in Fig 3A. The constitutive activity of mutated PDGFRα (mPDGFR) is inducible by doxycycline (DOX). Downstream of mPDGFR includes 3 canonical intracellular signalling pathways: MAPK, PI3K/AKT/mTOR and PLCγ/PKC pathways as well as regulatory mechanisms on PDGFRα. In addition, we included, STAT5, which was shown to be rather activated by oncogenic PDGFRα mutants [13]. SHP2, PI3K and PLCγ, marked with asterisks, are the targets of YF mutants. The basal activities of PTEN and PDK were represented by bPTEN and bPDK, respectively. Four signalling inhibitors, i.e. Wortmannin, U0126, GF109231X (GF109), and Gö6976, and their targets are also depicted. The phosphorylation state of five downstream signalling molecules, i.e. PDGFRα, STAT5, PLCγ, ERK1,2 and AKT were used as read-outs. There were 9 crosstalk interactions proposed in literature (numbered 1–9) that might be involved in PDGF signalling. Note that the phosphorylation of PKC substrates (PKC) was also included as a read-out in the additional experiment (Fig 4) and the read-out nodes with multiple inputs, i.e. PDGFRα, PLCγ, PKC and AKT are also optimised. [B] The quantified Western blot data from 9 experimentally-investigated conditions together with a negative control condition were applied for the modelling task. Experimental data were split into a training set (single perturbation plus positive and negative controls) and a validation dataset (combinatorial perturbations).