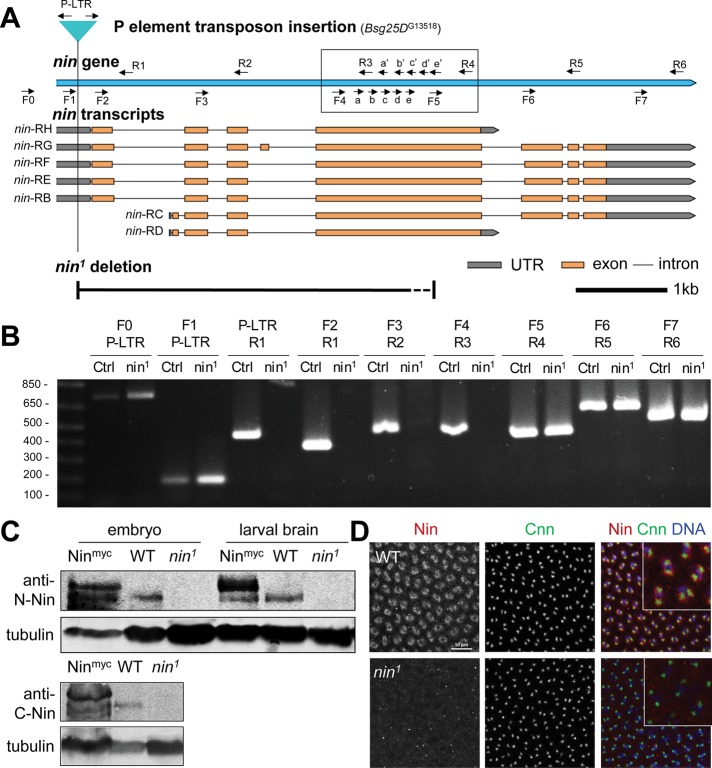
FIGURE 6:
nin1 is a deletion allele that disrupts nin expression. (A) Schematic view of Drosophila nin locus, transcripts, P element insertion (G13518) and nin1 deletion. The nin1 deletion allele was generated by mobilizing the P element transposon, Bsg25DG13518, located in the 5’ UTR. The primer pairs used for PCR screening are indicated (arrows). The primer pairs in the boxed region were used to narrow down the region deleted in nin1. The dotted line section of the deletion represents the region where the 3’ breakpoint of the nin1 deletion resides: somewhere between the “e” and “F5” primer sites. The scale bar is 1 kb. (B) Single adult fly PCR analysis of nin1, which shows a deletion of ∼3.5 kb. The PCR results for the primer pairs in the boxed region are shown in Supplemental Figure S4. Control (Ctrl) represents the original P element insertion stock that was used to generate the nin1 deletion allele. Sequences for the primers are listed in Supplemental Table S1. (C) Western blot analysis of nin1 embryo and larval brain lysates using an antibody against the N-terminal region of Nin, and in embryo lysates also using a C-terminal antibody. The Nin-myc transgene shows endogenous and myc-tagged Nin bands with an ∼150 kDa Mr, there is 6.4-fold increase in the tagged Nin expression compared with the endogenous Nin. Wild-type shows endogenous Nin bands, which are absent in nin1 lysates. α-tubulin serves as loading control. (D) Immunofluorescence staining of wild-type and nin1 embryos. Fixed embryos were stained with antibodies against endogenous Nin N-terminal region, and with Cnn antibody to mark centrosomes. The inset shows magnified view of Nin and Cnn in embryos.