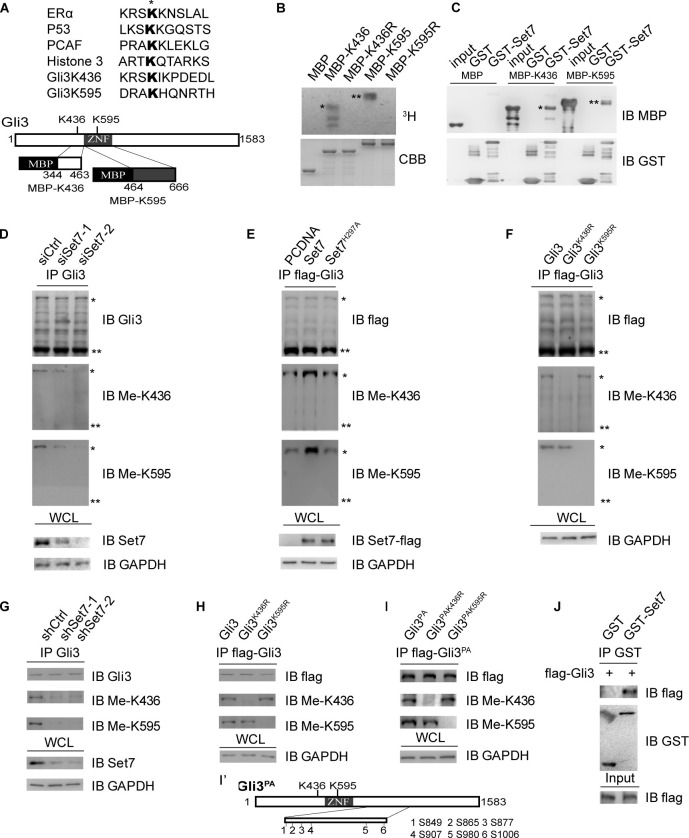
Figure 1. Set7 methylates Gli3 full-length in vivo and in vitro.
(A) Sequence alignment of the reported Set7 substrates with Gli3K436 and Gli3K595 (upper). Schematic representation of Gli3 protein and the truncated peptide used in in vitro methylation assay (B) and GST pull-down assay (C) (lower). (B) In vitro methylation assay with 3H-S-adenosine-methionine (3H-SAM), bacteria purified Set7 and MBP fusion protein. * and ** represent the MBP-K436 and MBP-K595 respectively. (C) GST pull-down assay using GST-Set7 and MBP tagged Gli3 truncated fragments described in (A). * and ** represent the MBP-K436 and MBP-K595 respectively. (D–F) Western blot of immunoprecipitates (top three panels) and lysates (bottom) from HEK293T cells expressing indicated siRNAs or proteins. * and ** represent the full-length and repressor forms of Gli3 respectively. (G–I) Western blot of immunoprecipitates (top three panels) and lysates (bottom) from NIH-3T3 cells stably expressing indicated shRNAs or proteins. (I’) Schematic representation of 6 PKA targeted serines which were mutated to nonphosphorylatable alanines in Gli3PA. (J) GST pull-down assay using GST-Set7 and flag-Gli3 in NIH-3T3 cells in the presence of Shh. Ctrl, Control. Me-K436, antibody anti methylated Gli3-K436; Me-K595, antibody anti methylated Gli3-K595. WCL, whole cell lysis. The protein level of Gli3 in (D–I) are normalized to the same.
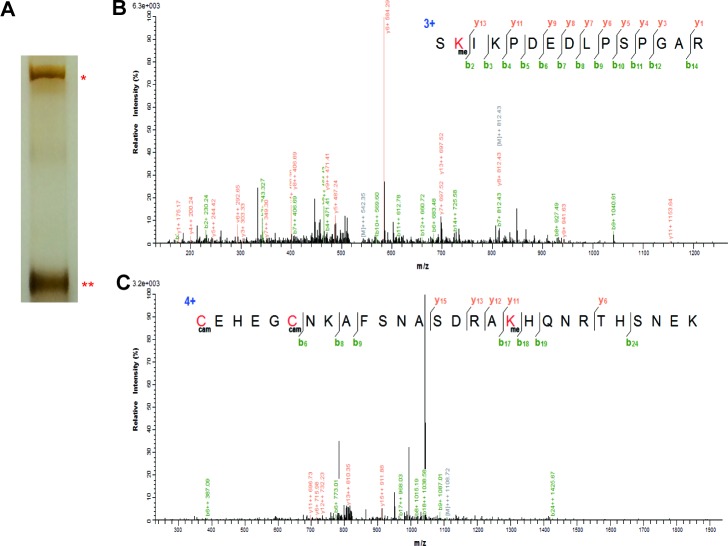
Figure 1—figure supplement 1. The mass spectrometry results show the methylation modification in the full-length Gli3 on K436 and K595.
Figure 1—figure supplement 2. Sequence alignment of K436 and K595 sites of Gli3 in different species Sequence alignment of the methylation sites K436.
Figure 1—figure supplement 3. The methylation antibodies can specifically recognize the mono methylated Gli3 peptides.
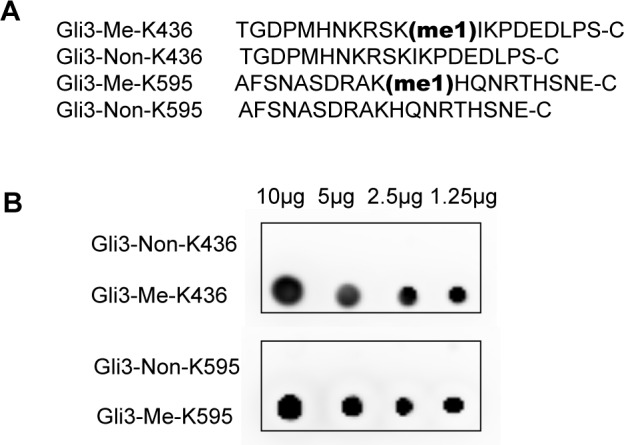
Figure 1—figure supplement 4. The methylation antibodies can specifically recognize the mono methylated Gli3 full length in embroynic lung Indicated tissues from mouse embryos (14.5 dpc) were isolated and lysed.