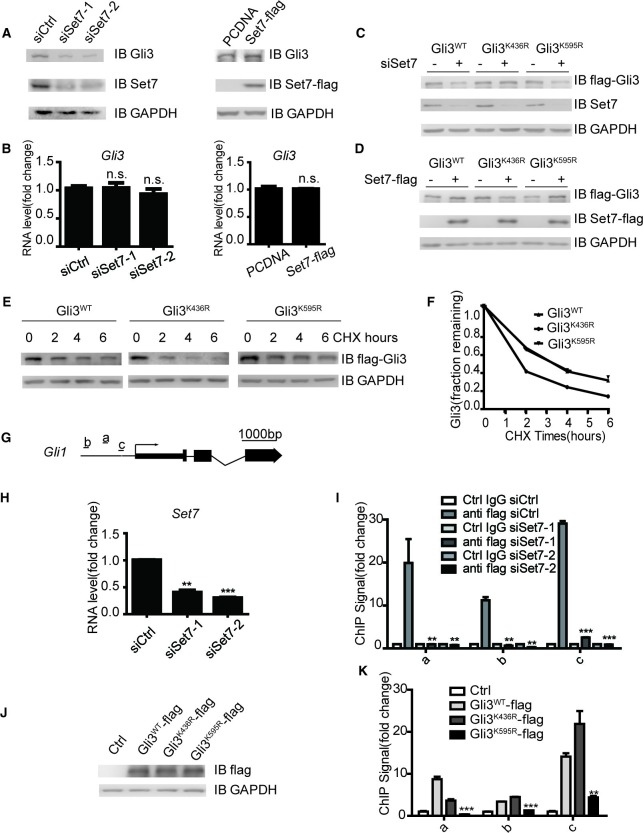
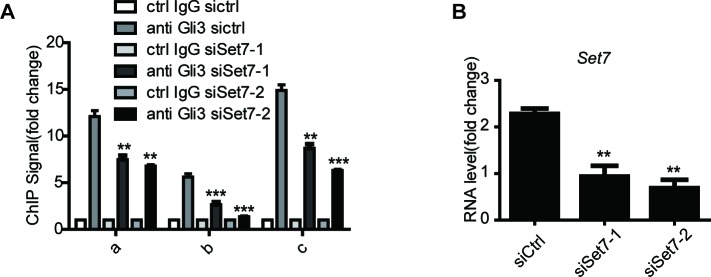
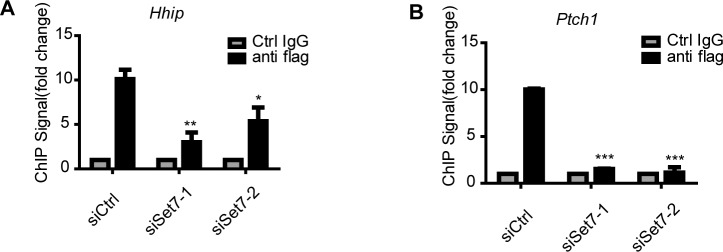
Figure 3. Set7 improved the stability and DNA binding ability respectably through K436 and K595 methylation.
(A and B) Western blot (A) and qPCR (B) of Gli3 in the condition of Set7 knockdown or overexpression in NIH-3T3 cells. n.s. p>0.05 versus siCtrl (left) or PCDNA (right) respectively. (C and D) Western blot of Gli3WT, Gli3K436R or Glli3K595R in the condition of Set7 knockdown or overexpression in NIH-3T3 cells. (E and F) Western blot (E) and statistic (F) of Gli3WT, Gli3K436R or Gli3K595R in NIH-3T3 cells treated with cycloheximide (CHX) for the indicated times. (G) Schematic representation of three Gli3 binding sites on the promoter regions of Gli1. (H) qPCR of Set7 in Gli3WT-flag stable cell lines after Set7 knockdown in (I). **p<0.01, ***p<0.00 versus siCtrl. (I) ChIP-qPCR analysis using Ctrl IgG (grey) or anti-flag antibody (black) in NIH-3T3 cells transfected with Ctrl or Set7 siRNAs. ChIP signal levels are represented as fold change of input chromatin. (J) Western blot of flag tagged Gli3WT and mutants in the stable cell lines used in (K). (K) ChIP-qPCR analysis using anti-flag antibody in NIH-3T3 cells expressing Gli3WT-flag, Gli3K436R-flag or Gli3K595R-flag. ChIP signal levels are represented as fold change of input chromatin. *p<0.05, **p<0.01, ***p<0.001, n.s. p>0.05 versus anti flag siCtrl (I) or Gli3WT-flag (K). Data are represented as mean ± SEM (n=3). All the qPCR results are normalized to GAPDH. In all the experiments, cells are treated with Shh. Ctrl, Control.