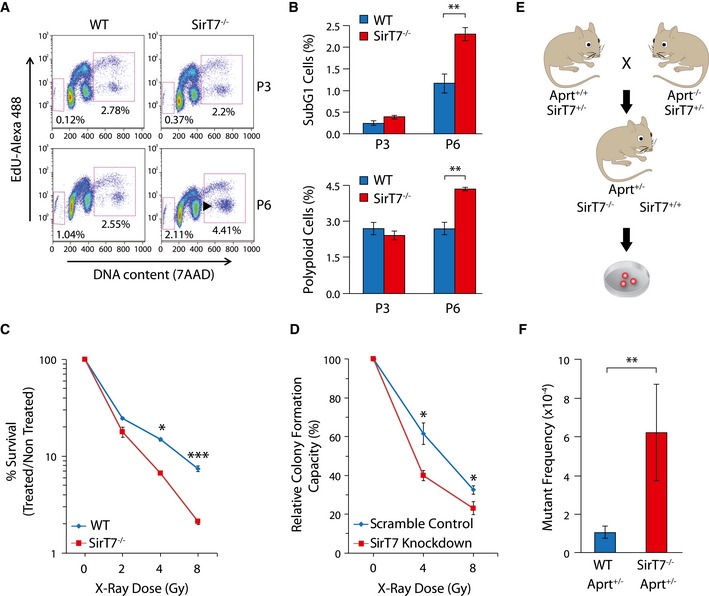
Figure 3. Increased genome instability in SirT7 −/− mice.
-
A, BDot plots of FACS cell cycle analyses of WT and SirT7 −/− MEFs in passages 3 (P3) and 6 (P6) using EdU incorporation and 7AAD (A). Percentages of cells in sub‐G1 (apoptotic cells, left square) and cells with DNA content above 4N (polyploid, right square). (B) Quantitation of experiment shown in (A) (mean ± SEM; three samples per genotype).
-
CSurvival curve for WT and SirT7 −/− thymocytes after X‐ray irradiation (IR) at the indicated doses. Cell death was quantified by FACS using Annexin V and 7AAD staining 18 h postinsult (mean ± SEM; three samples per genotype from one of two independent experiments).
-
DClonogenic assays in HT1080 cells transfected with scramble control or SirT7 knockdown and irradiated at the indicated X‐ray doses, then plated at low density. After 9 days, colonies were stained with crystal violet and counted (mean ± SEM; three independent cell lines per genotype).
-
ESchematic describing the mouse APRT loss of heterozygosity (LOH) assays. Aprt +/− mice were used to measure the in vivo somatic mutation frequency in WT and SirT7 −/− mice. Cells fully deficient for APRT are selected in culture by 2,6‐diaminopurine (DAP), an adenine analog that is converted to a toxic product by APRT enzymatic activity. Mutant frequency is proportional to the number of DAP‐resistant (DAP r) colonies.
-
FQuantitation of experiment shown in E (mean ± SEM; 9 WT and 3 SirT7 −/− samples).
