Abstract
Introduction
The genetic background of atopic dermatitis (AD) is complex, involves many genes and their participation varies in varied populations, and depends on the intensity and course of a disease. Changes in the nucleotide sequence of the FLG gene and a reduced number or a deficit of the functional product of processed profilaggrin can be one of risk factors for atopic dermatitis.
Aim
To determine the prevalence of R501X and 2282del4 mutations of the FLG gene in patients with AD.
Material and methods
The studied group included 60 patients with clinically diagnosed AD, and the control group included 61 healthy volunteers. The study protocol included collection of biological material for tests, DNA isolation and evaluation of its quality and quantity, and PCR amplification of the isolated genetic material.
Results
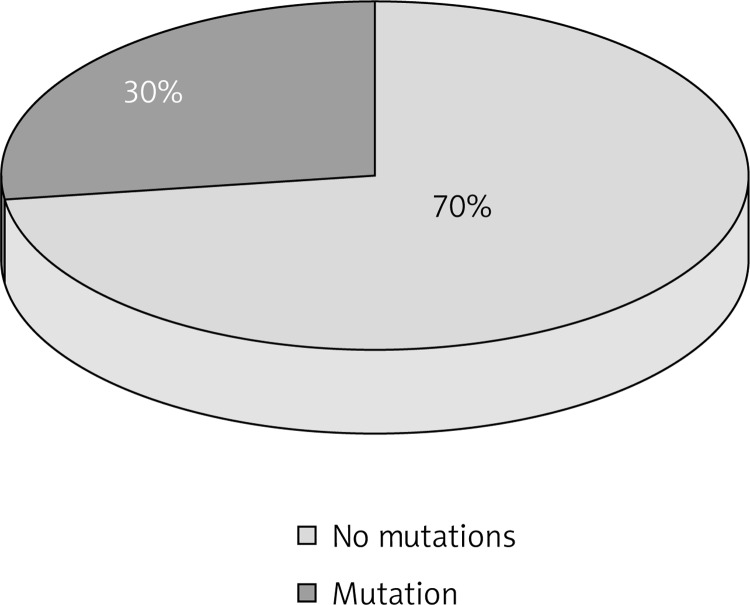
In the studied group, both changes in the nucleotide sequence of the FLG gene were detected and in the control group no tested mutations were detected. In 18 (30%) patients with AD, 22 mutations (4 heterozygous and 1 homozygous ones of R501X and 10 heterozygous and 7 homozygous ones of 2282del4) were detected.
Conclusions
A high rate of mutations of the FLG gene in patients with clinically diagnosed AD and pathologically dry skin was observed in the studied population. The 2282del4 mutation occurred more often than R501X.
Keywords: dry skin, atopic dermatitis, mutations, filaggrin
Introduction
Epidermal proteins, in particular filaggrin (filament aggregation protein), one of markers of terminally differentiating keratinocytes, play a significant role in the process of keratinization, formation and normal functioning of the epidermal protective layer. Abnormalities at different stages of its production and further decomposition are a cause of disturbances in the process of terminal cell differentiation, increased permeability of the epidermal barrier, resulting in dry skin that is a key diagnostic criterion of atopic dermatitis (AD). Apart from complex interactions between exogenous and immune factors, genetic conditions also play a role in the development of AD, and inheritance depends on many genes. However, genetic background of this dermatosis is complex, includes many genes and their participation in different populations varies, and also depends on the intensity and the course of this disease [1–5]. Among others, polymorphisms/mutations in a gene group forming the epidermal differentiation complex (EDC) are responsible for genetically conditioned damage to the dermal barrier in the course of AD. Changes in a nucleotide sequence of the FLG gene located on chromosome 1q21, namely a reduced number or a deficit of the functional product of processed profilaggrin can be one of risk factors for ichthyosis and simultaneously constitute one of risk factors for AD and moreover, they might also correlate with a phenotype and a clinical course of the disease [1, 2, 6–8]. Reduced expression of filaggrin due to structural changes in the FLG gene has been confirmed in pathologically changed and intact skin of patients with AD [9]. Two most frequent FLG mutations in European population, R501X and 2282del4, first described by Palmer in 2006, were studied, because both of them are highly predisposing to development of ichthyosis vulgaris as well as to AD.
Aim
The objective of the work was to determine the prevalence of R501X and 2282del4 mutations of the FLG gene in patients with AD.
Material and methods
The analysis of prevalence of R501X and 2282del4 mutations in the gene encoding filaggrin was performed in 121 adults.
The study protocol was approved Ethical Committee of Collegium Medicum in Bydgoszcz (KB 536/2010 dated 3 December 2010).
The studied group included 60 (49.59%) patients with AD and pathologically dry skin aged between 18 and 61 years (mean age: 29.92 years).
The control group included 61 (50.41%) healthy volunteers without symptoms of dry skin in whom dermatologic and allergic conditions were excluded, aged between 18 and 76 years (mean age: 42.41 years).
The material for analysis included whole peripheral blood collected from study participants by qualified medical personnel into 2-ml tubes with EDTA (1 mg/ml). Identical composition of the reacting mixture and individually chosen primers were used for each reaction and for both mutations.
FLG R501X 5’ – ACA GCC TGA CTC TGC CCA TG – 3’ (forward)
FLG R501X 5’ – GCA CTT CTG GAT CCT GAC TG – 3’ (reverse)
FLG 2282del4 5’ – TCC CGC CAC CAG CTC C – 3’ (forward)
FLG 2282del4 5’ – TG GCT CTG CTG ATG GTG A – 3’ (reverse).
The study protocol included collection of biological material for tests, DNA isolation and evaluation of its quality and quantity, and then PCR amplification of the isolated genetic material was performed. After multiplication, the PCR products were digested with a restrictive enzyme and results were visualized in the PAA gel.
Statistical analysis
The statistical analysis was performed with the statistical software Statistica 10.0 by StatSoft®. Quantitative variables (subjects’ age) were described with the following statistics: number (N), arithmetic mean (M), standard deviation (SD), median (Me), minimum (Min.) and maximum (Max.). The analysis of a qualitative variable correlation was performed with the χ2 test. For 2 x 2 tables and theoretical numbers below 5, the Yates’ correction was used. The test likelihood at p < 0.05 was considered to be statistically significant and marked with “*”. The analysis was also supplemented with calculated odd ratios (OR).
Results
In the studied group both changes in a nucleotide sequence of the FLG gene (Figure 1) were detected and in the control group, no tested mutations were detected.
Figure 1.
Prevalence of the FLG gene mutations in patients with AD
In 18 (30%) patients with AD, 22 mutations (4 heterozygous and 1 homozygous ones of R501X and 10 heterozygous and 7 homozygous ones of 2282del4) were detected (Figure 1).
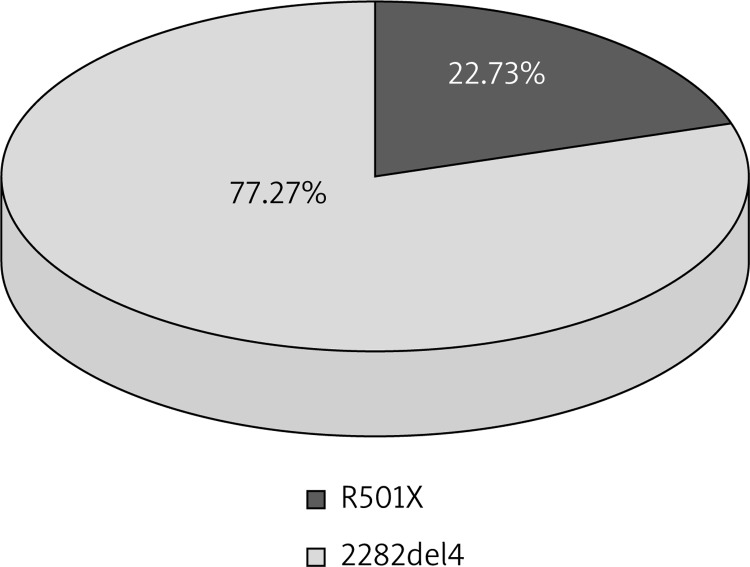
In 4 patients at the same time both genetic variants were present (4 heterozygous ones of R501X and 2 heterozygous and 2 homozygous ones of 2282del4) (Table 1, Figure 2).
Table 1.
Patients with AD and pathologically dry skin in whom R501X and/or 2282del4 mutations were observed
| No. | Phenotype | Age | Gender | R501X | 2282del4 | ||
|---|---|---|---|---|---|---|---|
| Heterozygotes | Homozygotes | Heterozygotes | Homozygotes | ||||
| 1 | m | 28 | F | + | |||
| 2 | mr | 24 | F | + | + | ||
| 3 | s | 21 | F | + | + | ||
| 4 | s | 35 | F | + | |||
| 5 | m | 22 | F | + | |||
| 6 | m | 28 | M | + | |||
| 7 | m | 40 | F | + | |||
| 8 | m | 26 | F | + | |||
| 9 | m | 27 | F | + | |||
| 10 | m | 24 | M | + | |||
| 11 | mr | 18 | F | + | + | ||
| 12 | s | 56 | M | + | |||
| 13 | s | 56 | M | + | |||
| 14 | s | 37 | F | + | |||
| 15 | s | 51 | F | + | |||
| 16 | s | 56 | F | + | + | ||
| 17 | s | 23 | F | + | |||
| 18 | m | 29 | F | + | |||
Phenotype: m – mild, mr – moderate, s – severe, gender: F – female, M – male.
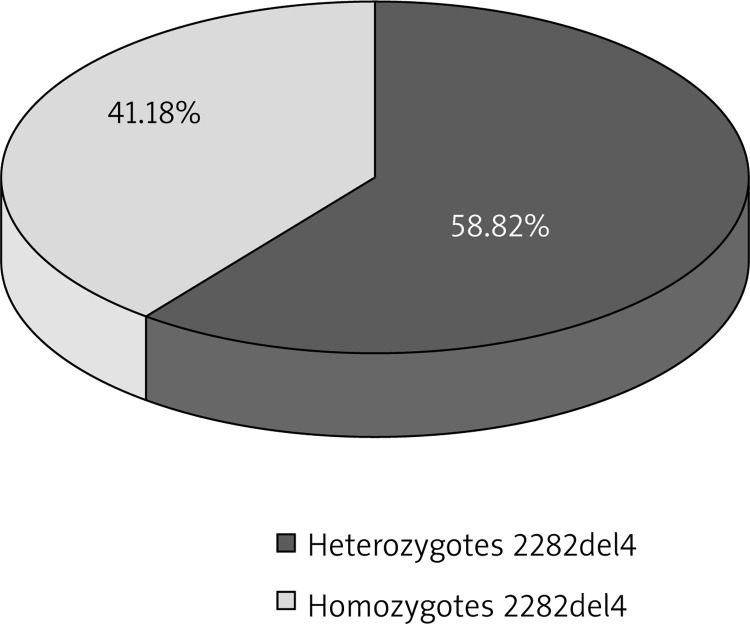
Figure 2.
Prevalence of R501X and 2282del4 mutations in patients with AD
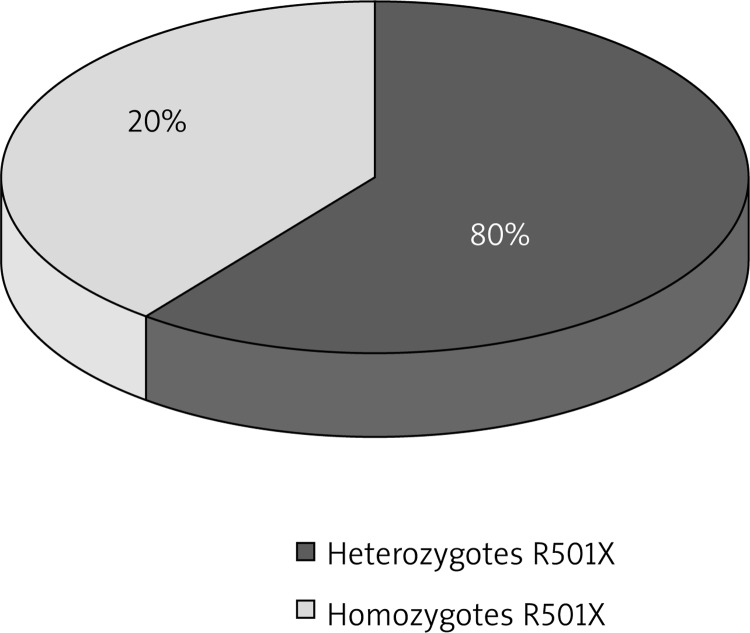
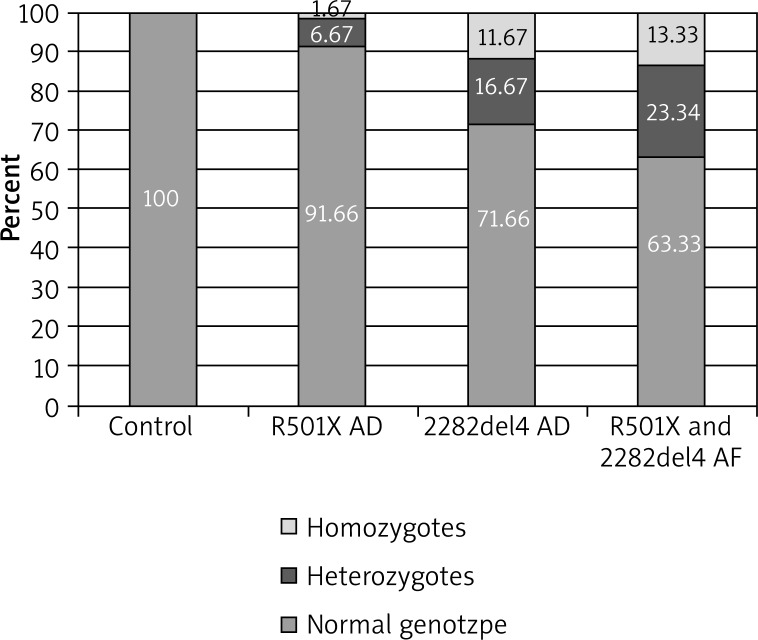
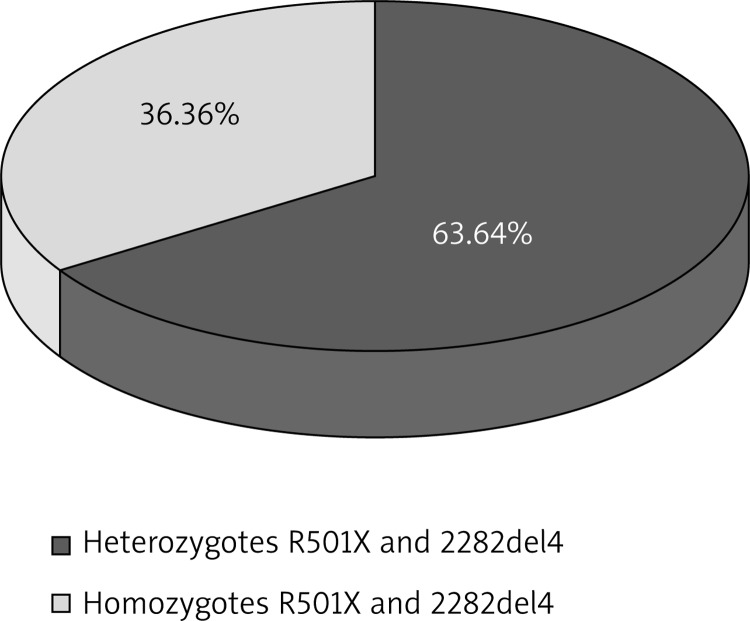
In the case of the R501X mutation, 4 (6.67%) heterozygotes and 1 (1.67%) homozygote were observed, and differences with regard to the total prevalence of heterozygotes and homozygotes for nonsense mutation between the AD group and the control group were close to the statistical significance (p = 0.0706). In 61 subjects from the control group (100.00%) and 55 from the studied group (91.66%), a normal genotype with regard to this mutation was demonstrated (Table 2, Figures 3, 4).
Table 2.
Correlation between the prevalence of the R501X mutation and AD development
| R501X | Control group | Studied group | OR* | P-value | ||
|---|---|---|---|---|---|---|
| 1 | 2 | 1–2 | ||||
| N | % | N | % | |||
| Total | 61 | 100.0 | 60 | 100.0 | 5.56 | 0.0706 |
| Normal genotype | 61 | 100.0 | 55 | 91.6 | ||
| Heterozygotes | 0 | 0.0 | 4 | 6.6 | ||
| Homozygotes | 0 | 0.0 | 1 | 1.6 | ||
Figure 3.
Prevalence of R501X heterozygotes and homozygotes in patients with AD
Figure 4.
Prevalence of R501X and 2282del4 mutations in the control group and in patients with AD
When analyzing the prevalence of the 2282del4 mutation, the presence of 10 (16.67%) heterozygotes and 7 (11.67%) homozygotes was observed. A performed statistical analysis revealed significant differences between the total prevalence of heterozygous and homozygous genotypes as compared to the control group (p < 0.0001). A normal genotype with regard to this mutation was present in 43 subjects in the studied group (71.66%) and 61 subjects in the control group (100.00%) (Table 3, Figures 4, 5).
Table 3.
Correlation between the prevalence of the 2282del4 mutation and AD development
| 2282del4 | Control group | Studied group | OR* | P-value | ||
|---|---|---|---|---|---|---|
| 1 | 2 | 1–2 | ||||
| N | % | N | % | |||
| Total | 61 | 100.0 | 60 | 100.0 | 24.15 | < 0.0001* |
| Normal genotype | 61 | 100.0 | 43 | 71.6 | ||
| Heterozygotes | 0 | 0.0 | 10 | 16.6 | ||
| Homozygotes | 0 | 0.0 | 7 | 11.6 | ||
Figure 5.
Prevalence of 2282del4 heterozygotes and homozygotes in patients with AD
When considering the total prevalence of the R501X substitution and the 2282del4 deletion in the studied group, 14 (23.34%) heterozygotes and 8 (13.33%) homozygotes were observed, and a statistical analysis performed also indicated significant differences between the total prevalence of heterozygous and homozygous genotypes as compared to the control group (p < 0.0001) (Table 4, Figures 4, 6).
Table 4.
Correlation between the prevalence of the FLG mutation and AD development
| R501X and 2282del4 | Control group | Studied group | OR* | P-value | ||
|---|---|---|---|---|---|---|
| 1 | 2 | 1–2 | ||||
| N | % | N | % | |||
| Total | 61 | 100.0 | 60 | 100.0 | 35.32 | < 0.0001* |
| Normal genotype | 61 | 100.0 | 38 | 63.3 | ||
| Heterozygotes | 0 | 0.0 | 14 | 23.3 | ||
| Homozygotes | 0 | 0.0 | 8 | 13.3 | ||
Figure 6.
Total prevalence of heterozygotes and homozygotes in patients with AD
Based on the results obtained, it was concluded that the presence of the deletion increases the likelihood of AD 24 times (OR = 24.15), and its combination with the substitution increases this risk more than 35 times (OR = 35.32).
Discussion
According to the authors, FLG gene mutations predispose to ichthyosis and AD that often coexists with ichthyosis. Our own observations confirm a semidominant inheritance of AD determined by Palmer (high penetration in FLG-null homozygotes/complex heterozygotes, low in heterozygotes) [1, 2, 7, 9]. A molecular analysis of the genetic material collected from patients confirmed their participation in epidermal barrier dysfunctions. Probably the first dermal lesions are located in the areas where the stratum corneum, namely the protective barrier is the thinnest, what additionally makes it easier for irritants and allergens to pass [10]. Kezic et al. conclude that abnormalities in a process of filaggrin formation and its further decomposition are also a cause of dry skin [11]. On the other hand, Weidinger et al. demonstrated a correlation between FLG gene mutations and exogenous AD accompanied by increased IgE levels and positive spot tests [12]. However, the authors did not observe any significant correlations between these changes and endogenous AD, whereas Marenholtz et al. indicated a growing predisposition for EADS and IADS as well [13, 14]. According to Stemmler et al., R501X and 2282del4 mutations are present in 10.3% of patients in whom AD developed at the age above 2 years, and in 12.1% of patients in whom this condition developed earlier. The presence of at least one mutated allele was confirmed in 21.3% of patients and as a result it was possible to draw a hypothesis assuming a correlation between FLG gene mutations and an early start of AD [15]. However, neither Weidinger et al. nor Lesiak et al. demonstrated such a correlation [2, 14]. In our work we did not perform a detailed assessment of the correlation between a genetic defect in the FLG gene and an early start of AD because information about when the disease had started was only and exclusively taken from a medical history collected earlier, as subjects below 18 years old were excluded. Both mutations predispose to the development of atopic dermatitis with a moderate or severe, long-term and recurrent course [16, 17]. When verifying a correlation between these mutations and disease severity Lesiak et al. indicated that the 2282del4 mutation increases the risk of a moderate and severe form of AD more than two-fold what complies with the Morar's reports [2, 18]. Additionally, Ekelund et al. noticed that FLG-null variants (homozygous mutations) are more prevalent in the case of a severe AD phenotype [19]. Results of studies by Marenholz et al. and Weidinger et al. indicate a significant correlation between nonsense mutations in the filaggrin gene and AD, and this relationship regards approximately 30% of patients with this disease [13, 14]. An identical result, approximately 30%, was also obtained in our own study; however, for both mutations together. Thanks to the analysis of correlations between said mutations and a phenotype of atopic dermatitis, the Sandilands’ team was able to present a thesis that R501X as well as 2282del4 are present first of all in patients in whom dominant features of the clinical manifestation include dry skin, keratosis pilaris and increased crease formation on the palmar surfaces of the hands [20]. In our own study, a correlation with dry skin was first of all demonstrated, what probably is a result of the fact that symptoms of dry skin were present in all patients with AD. Lack of a correlation between studied mutations and ichthyosis is a result of patients’ selection as the study did not include patients with this genodermatosis.
Conclusions
A high rate of mutations of the FLG gene in patients with clinically diagnosed AD and pathologically dry skin was observed in the studied population. The 2282del4 mutation was present more often than R501X in the studied population.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Hawro T, Sysa-Jędrzejowska A, Narbutt J. The role of filaggrin gene mutations in pathogenesis of atopic dermatitis. Literature review. Postep Derm Alergol. 2008;25:12–5. [Google Scholar]
- 2.Lesiak A, Przybyłowska K, Zakrzewski M. Mutacje R501X i 2282del4 w genie filagryny a atopowe zapalenie skóry. Alerg Astma Immun. 2010;15:162–9. [Google Scholar]
- 3.Woźniak M, Zegarska B, Romańska-Gocka K. Sucha skóra – problem kosmetyczny i dermatologiczny związany z negatywnym wpływem środowiska zewnętrznego. In: Bartuzi Z, editor. Interdyscyplinarny wymiar nauk o zdrowiu. Bydgoszcz: UMK CM; 2007. pp. 488–92. [Google Scholar]
- 4.Nedoszytko B, Sokołowska-Wojdyło M, Ruckemann-Dziurdzińska K, et al. Chemokines and cytokines network in the pathogenesis of the inflammatory skin diseases: atopic dermatitis, psoriasis and skin mastocytosis. Postep Derm Alergol. 2014;31:84–91. doi: 10.5114/pdia.2014.40920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rutkowski K, Sowa P, Rutkowska-Talipska J, et al. Allergic diseases: the price of civilisational progress. Postep Derm Alergol. 2014;312:77–83. doi: 10.5114/pdia.2014.40936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segre JA. Epidermal differentiation complex yields a secret: mutations in the cornification protein filaggrin underlie ichtyosis vulgaris. J Invest Dermatol. 2006;126:1202–4. doi: 10.1038/sj.jid.5700367. [DOI] [PubMed] [Google Scholar]
- 7.Smith FJ, Irvine AD, Terron-Kwiatkowski A. Loss of function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 2006;38:337–42. doi: 10.1038/ng1743. [DOI] [PubMed] [Google Scholar]
- 8.Wu Z, Hansmann B, Meyer-Hoffert U. Molecular identification and expression analysis of filaggrin-2, a member of the S100 fused-type protein family. PLoS One. 2009;4:e5227. doi: 10.1371/journal.pone.0005227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palmer CN, Irvine AD, Terron-Kwiatkowski A. Common loss of function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–6. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 10.Kruszewski J. Genetyczne uwarunkowanie atopii. Postep Derm Alergol. 2003;20:58–64. [Google Scholar]
- 11.Kezic S, Kemperman PM, Koster ES. Loss-of-function mutations in the filaggrin gene lead to reduced level of natural moisturizing factor in the stratum corneum. J Invest Dermatol. 2008;128:2117–9. doi: 10.1038/jid.2008.29. [DOI] [PubMed] [Google Scholar]
- 12.Weidinger S, Rodriguez E, Stahl C. Filaggrin mutations strongly predispose to early-onset and extrinsic atopic dermatitis. J Dermatol. 2007;127:724–6. doi: 10.1038/sj.jid.5700630. [DOI] [PubMed] [Google Scholar]
- 13.Marenholz I, Nickel R, Rüschendorf F. Filaggrin loss of function mutations predispose to phenotypes involved in the atopic march. Allergy Clin Immunol. 2006;118:866–71. doi: 10.1016/j.jaci.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 14.Weidinger S, Illig T, Baurecht H. Loss of function variations within the filaggrin gene predispose for atopic dermatitis with allergic sensitizations. J Allergy Clin Immunol. 2006;118:214–9. doi: 10.1016/j.jaci.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Stemmler S, Parwez Q, Petrasch-Parwez E. Two common loss-of-function mutations within the filaggrin gene predispose for early onset of atopic dermatitis. J Invest Dermatol. 2007;127:722–4. doi: 10.1038/sj.jid.5700579. [DOI] [PubMed] [Google Scholar]
- 16.Leung DY. Our evolving understanding of the functional role of filaggrin in atopic dermatitis. J Allergy Clin Immunol. 2009;124:494–5. doi: 10.1016/j.jaci.2009.07.041. [DOI] [PubMed] [Google Scholar]
- 17.Van den Oord RA, Sheikh A. Filaggrin gene defects and risk of developing allergic sensitization and allergic disorders: systematic review and meta-analysis. BMJ. 2009;339:b2433. doi: 10.1136/bmj.b2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morar N, Cookson WO, Harper JI. Filaggrin mutation in children with severe atopic dermatitis. J Invest Dermatol. 2007;127:1667–72. doi: 10.1038/sj.jid.5700739. [DOI] [PubMed] [Google Scholar]
- 19.Ekelund E, Lieden A, Link J. Loss-of-function variants of the filaggrin gene are associated with atopic eczema and associated phenotypes in Swedish families. Acta Derm Venereol. 2008;88:15–9. doi: 10.2340/00015555-0383. [DOI] [PubMed] [Google Scholar]
- 20.Sandilands A, O'Reagan GM, Liao H. Prevalent and rare mutations in the gene encoding filaggrin cause ichthyosis vulgaris and predispose individuals to atopic dermatitis. J Invest Dermatol. 2006;126:1770–5. doi: 10.1038/sj.jid.5700459. [DOI] [PubMed] [Google Scholar]