Figure 4. NUAK1 was identified as a target gene of miR-203.
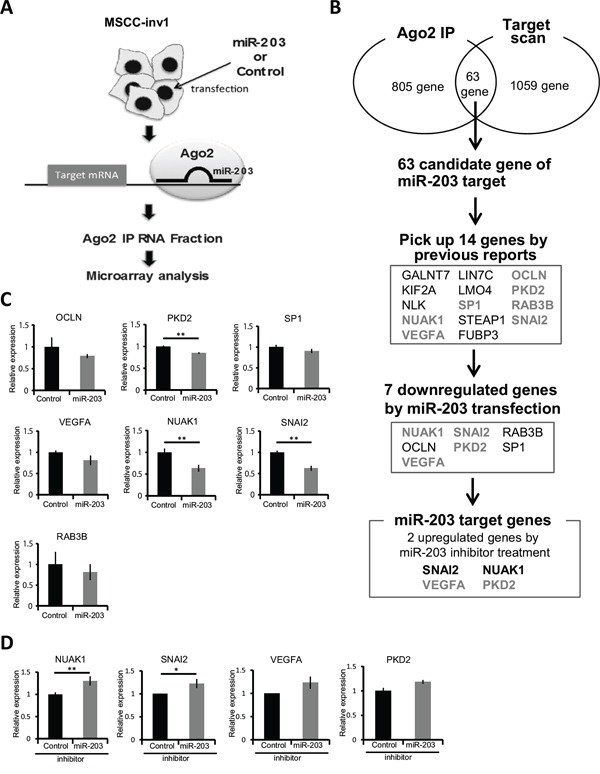
A. Schematic representation of the process of finding the target gene of miR-203. Mature miR-203 or control miRNA was transfected into MSCC1-inv1 cells. Then, miRNAs and target mRNAs were immunoprecipitated with anti-Ago2 antibody. Isolated mRNAs were analyzed by microarray. B. In MSCC-inv1 cells with ectopic miR-203 expression, 805 genes were enriched at least 2-fold over controls. By using Target Scan algorithm analysis (http://www.targetscan.org/), 1059 genes were predicted as target genes of miR-203. Sixty-three genes overlapped in the two analyses. Among those 63 genes, 14 were picked up by previous reports on the involvement of cancer, invasion, and EMT (Supplemental Table 1) and the result of Ago2-IP microarray. We then screened these genes by mature miR-203 transfection and miR-203 inhibitor transfection. Finally, we identified NUAK1 and SNAI2 as target genes of miR-203. C. The graph shows the expression of candidate genes of miR-203 (OCLN, PKD2, SP1, RAB3B, VEGFA, NUAK1, and SNAI2) in control- or mature miR-203-transfected MSCC-inv1 cells. The graph shows the expression of these mRNAs (mRNA/GAPDH). The results are presented as means ± SD. **P < 0.01. D. The graph shows the expression of NUAK1, SNAI2, VEGFA, and PKD2 in control- or miR-203 inhibitor-transfected HSC2 cells. The graph shows the expression of these mRNAs (mRNA/GAPDH). The results are presented as means ± SD. **P < 0.01, *P < 0.05.
