Figure 7. Downregulation of miR-203 by hypermethylation.
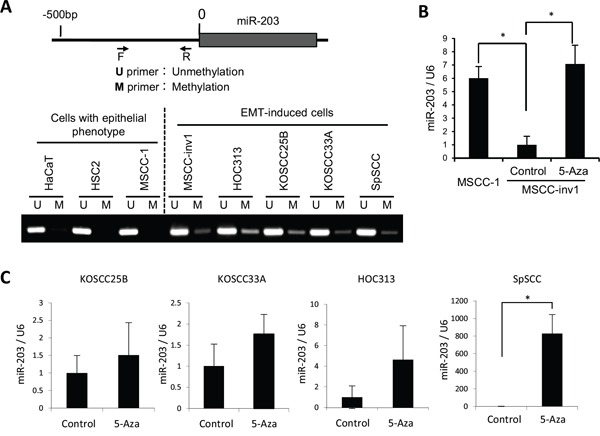
A. Analyzed regions of miR-203 are shown in the upper panel. The genomic DNA was isolated, treated with bisulfite, and then subjected to MS-PCR in HaCaT and HNSCC cells (HSC2, MSCC-1, MSCC-inv1, HOC313, KOSCC25B, KOSCC33A, and SpSCC). The primer sets that were specific for the methylated CpG (M) and the unmethylated CpG (U) were used for MS-PCR. The lower panel shows methylation status of miR-203 in cells with epithelial phenotype (HaCaT, HSC2, and MSCC-1) and EMT-induced cells (MSCC-inv1, HOC313, KOSCC25B, KOSCC33A, and SpSCC). B. To demethylate, MSCC-inv1 cells were treated with 5-aza-dC. Expression of miR-203/U6 was examined by real-time PCR, in comparison with MSCC-1 and MSCC-inv1 cells without 5-aza-dC treatment. The results are presented as means ± SD. *P < 0.05. C. To demethylate, KOSCC25B, KOSCC33A, HOC313, and SpSCC cells were treated with 5-aza-dC. Expression of miR-203/U6 was examined by real-time PCR. The results are presented as means ± SD. *P < 0.05.
