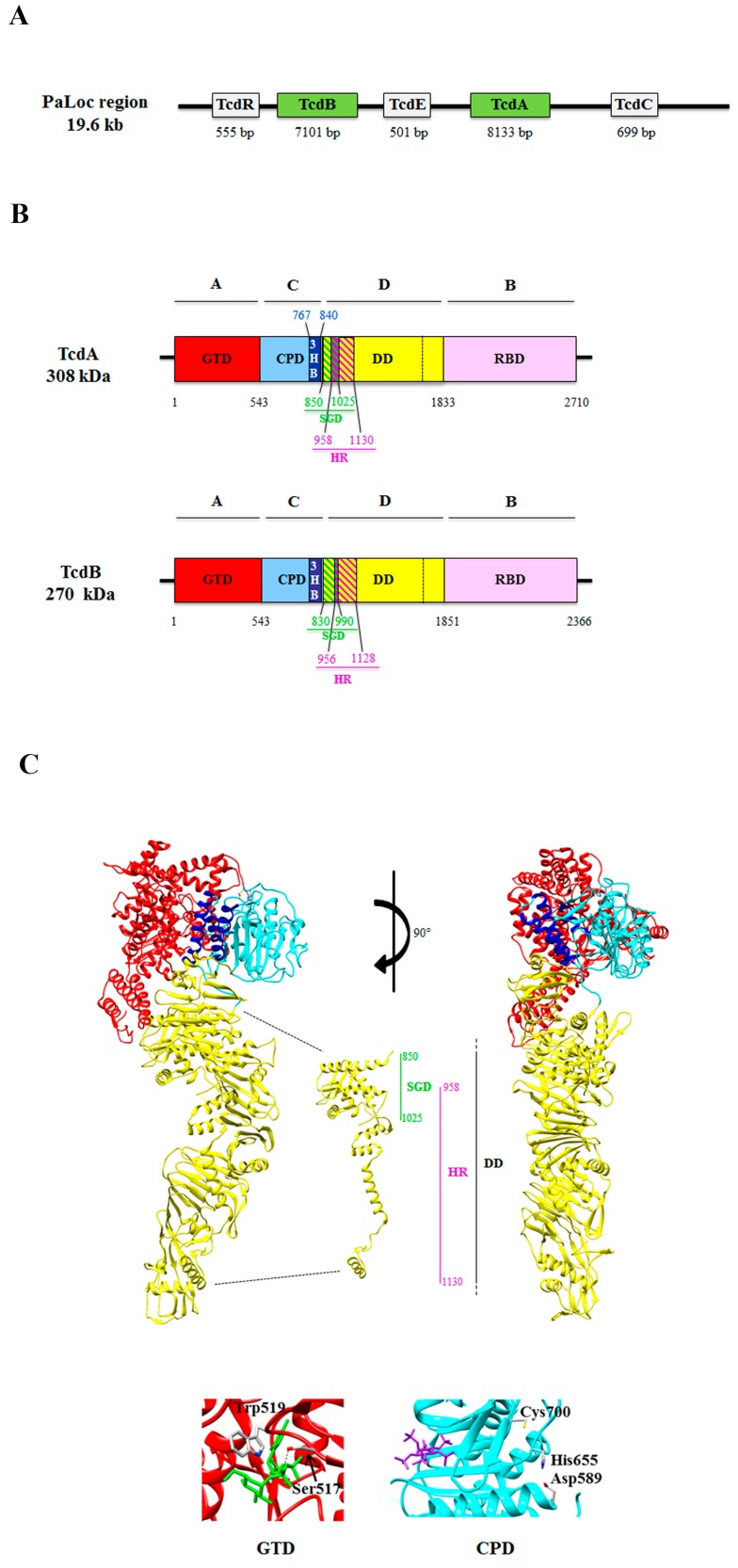
Figure 1.
(A) Schematic representation of the PaLoc region containing the following genes: tcdR, tcdB, tcdE, tcdC, and tcdA. Below each gene, the number of base pairs (bp) is indicated. (B) Schematic representation of the multi-modular domain structure described as the ABCD model (A: biological activity; B: binding; C: cutting; D: delivery) domain structure of TcdA and TcdB. The two toxins are composed of: the A domain, corresponding to the N-terminal glucosyltransferase domain (GTD) (red) [32,33]; the C domain, corresponding to the cysteine protease domain (CPD) (cyan) and the three-helix bundle domain (3HB) (blue) identified in TcdA [34], and possibly present also in TcdB; the D domain, which corresponds to the delivery hydrophobic domain (DD) (yellow), containing the small globular domain (SGD) in TcdA (green and yellow diagonal lines) [34] that corresponds to the minimal pore forming region (MPFR) in TcdB (green and yellow diagonal lines) [35], and overlapping only partially (purple and green diagonal lines) to the hydrophobic region (HR) of TcdA [36] and TcdB [35] (purple and yellow diagonal lines); the B domain, corresponding to the receptor binding domain (RBD) (pink) [34,37]. (C) Top: two three-dimensional structures of TcdA are shown, rotated 90° each other (red: GTD; cyan: CPD; blue: 3HB; yellow: DD; pink: RBD), with a detail of the N-terminal region of the D domain showing the SGD and HR; right bottom: detail of the loop containing the amino acid residues 516–522 of the GTD, in which the conserved Ser517 residue forms a hydrogen bond with the phosphate group present in the UDP-glucose, and the conserved Trp519 forms a hydrogen bond with the glycosidic oxygen (PDB: 3SRZ; [18]); left bottom: detail of the catalytic triad Asp589-Hys655-Cys700 of the CPD, which is responsible for the proteolytic cleavage of the toxin dependent upon inositol hexakisphosphate (Insp6) (purple) binding (PDB: 3HO6; [38]). The three-dimensional structures were drawn with the UCSF-Chimera package [39]. For details, see text.