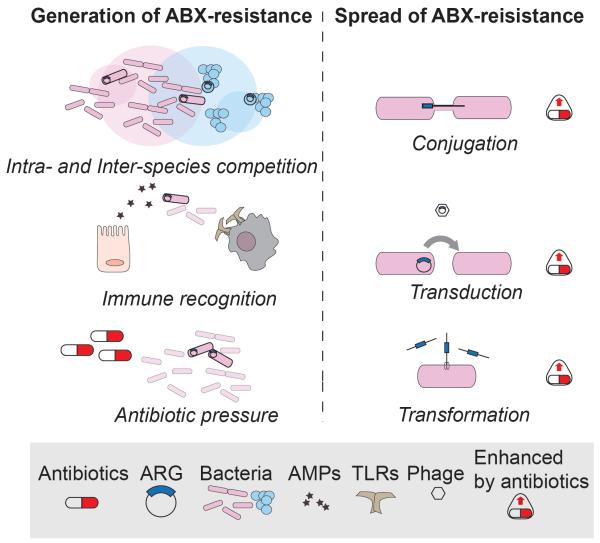
Figure 3. Generation and Spread of Antibiotic Resistance.
Left panel: antibiotic resistance is generated by mutations that can be induced by several driving forces. From the top: competition of bacteria (inter- or intra- species, here depicted as intersections between circles representing population niches) mainly mediated by bacteriocins, induces a selective pressure that favors development of resistance. Some TLRs or host-derived antimicrobial peptides (AMPs) target bacterial molecules that can undergo mutations and provide resistance to clinically-relevant antibiotics. Exogenous antibiotic pressure through medical or industrial practices (e.g.. antibiotic use in livestock) promotes the generation and selection of resistant strains that can rapidly diffuse. Right panel: antibiotic resistance genes can be exchanged among bacteria also of different species (not shown) through horizontal gene transfer, i.e. conjugation, transduction or transformation. Notably, these three mechanisms are all enhanced upon antibiotic exposure, resulting in a faster and more efficient spread of ARGs in the gut as well as in the environment.