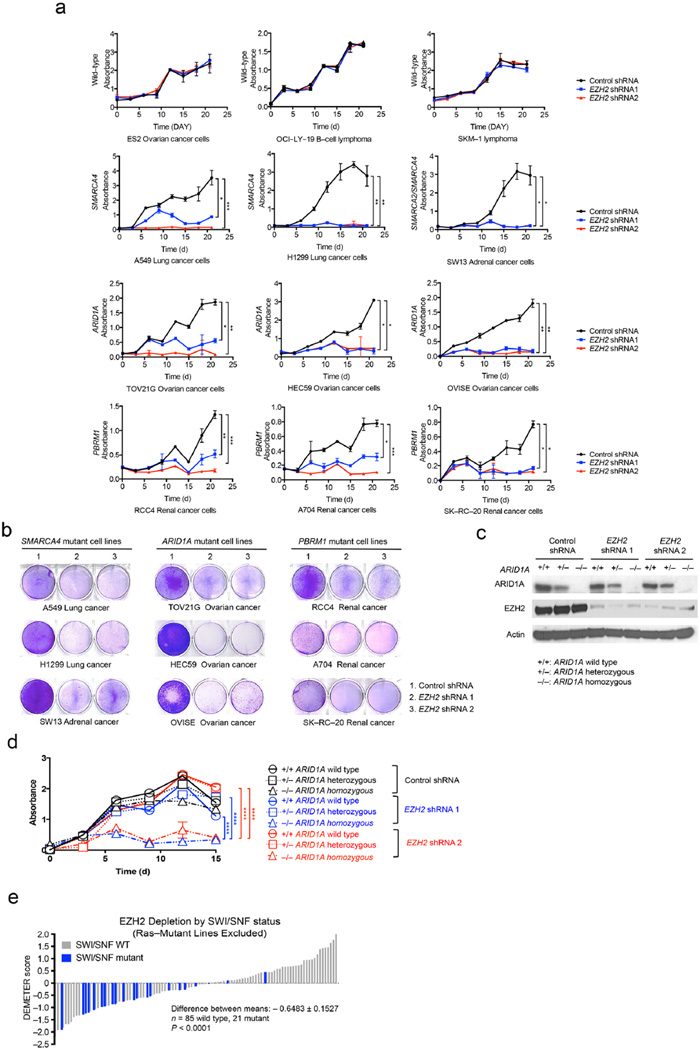
Figure 1. SWI/SNF mutant cancer cells require EZH2.
SWI/SNF mutant and wild–type cell lines were transduced with either control or EZH2–targeting shRNAs. Proliferation was assessed by MTT assay. (a) Proliferation curves of SWI/SNF wild–type cell lines (ES2, OCI–LY–19, and SKM–1) and mutant cancer cell lines including SMARCA4 mutant (A549, H1299, and SW13), ARID1A mutant (TOV21G, HEC59, and OVISE), and PBRM1 mutant (RCC4, A704, and SK–RC–20). Error bars indicate means ± s.d. (n = 3). (Ordinary one–way ANOVA, *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001) (b) Colony formation assays. Cells transduced with either control or EZH2 shRNAs were seeded at low density in standard 6–well plates. Colonies were visualized by crystal violet staining. The assays are representative of replicates of three independent experiments. (c) Western blot analysis of HCT116 isogenic cells (ARID1A wild-type parental, +/+; ARID1A heterozygous. +/−; or ARID1A homozygous deficient, −/−) after inducing with control or EZH2 shRNA. Actin was used as a loading control. (d). MTT proliferation curves of ARID1A isogenic cell lines. Error bars indicate means ± s.d. (Ordinary one–way ANOVA, *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (e) Distribution of EZH2 shRNA dependency based on SWI/SNF mutational status. Each bar represents one cell line; cell lines with homozygous inactivating SWI/SNF mutations are indicated in blue. Lower DEMETER scores indicates more dependency on EZH2. The statistical difference of the EZH2 dependency between SWI/SNF mutant and wild–type cells was calculated using an unpaired, two–sample Welch’s t–test.